Spatial transcriptome (VisiumHD)
We demonstrate a noise reduction with RECODE for spatial transcriptome data (FISH based). We use spatial transcriptome data of 10X Visium HD, Visium HD Spatial Gene Expression Library, Mouse Embryo (FFPE). The dataset is available from 10X datasets.
We use scanpy to read/write data. Import numpy and scanpy in addlition to screcode.
[1]:
import scanpy as sc
import numpy as np
import screcode
import warnings
warnings.simplefilter('ignore')
import matplotlib.pyplot as plt
import pandas as pd
Read in the count matrix into an AnnData object.
[2]:
INPUT_DIR = 'data/Visium_HD_Mouse_Embryo/Visium_HD_Mouse_Embryo_binned_outputs/binned_outputs/square_008um'
INPUT_FILE = "filtered_feature_bc_matrix.h5"
Raw_key = "count"
adata = sc.read_10x_h5("%s/%s" % (INPUT_DIR,INPUT_FILE))
df_spatial = pd.read_parquet("%s/spatial/tissue_positions.parquet" % (INPUT_DIR))
adata.obs = pd.concat([adata.obs,df_spatial.set_index("barcode").loc[adata.obs.index]],axis=1)
adata.var_names_make_unique()
adata = adata[:,np.sum(adata.X,axis=0)>0]
adata = adata[np.sum(adata.X,axis=1)>0]
adata.layers["Raw"] = adata.X.toarray()
adata
[2]:
AnnData object with n_obs × n_vars = 343983 × 19040
obs: 'in_tissue', 'array_row', 'array_col', 'pxl_row_in_fullres', 'pxl_col_in_fullres'
var: 'gene_ids', 'feature_types', 'genome'
layers: 'Raw'
Apply RECODE
Apply RECODE to the count matrix (without using spatial coordinates).
[3]:
import screcode
recode = screcode.RECODE(downsampling_rate=0.01)
adata = recode.fit_transform(adata)
start RECODE for scRNA-seq data
end RECODE for scRNA-seq
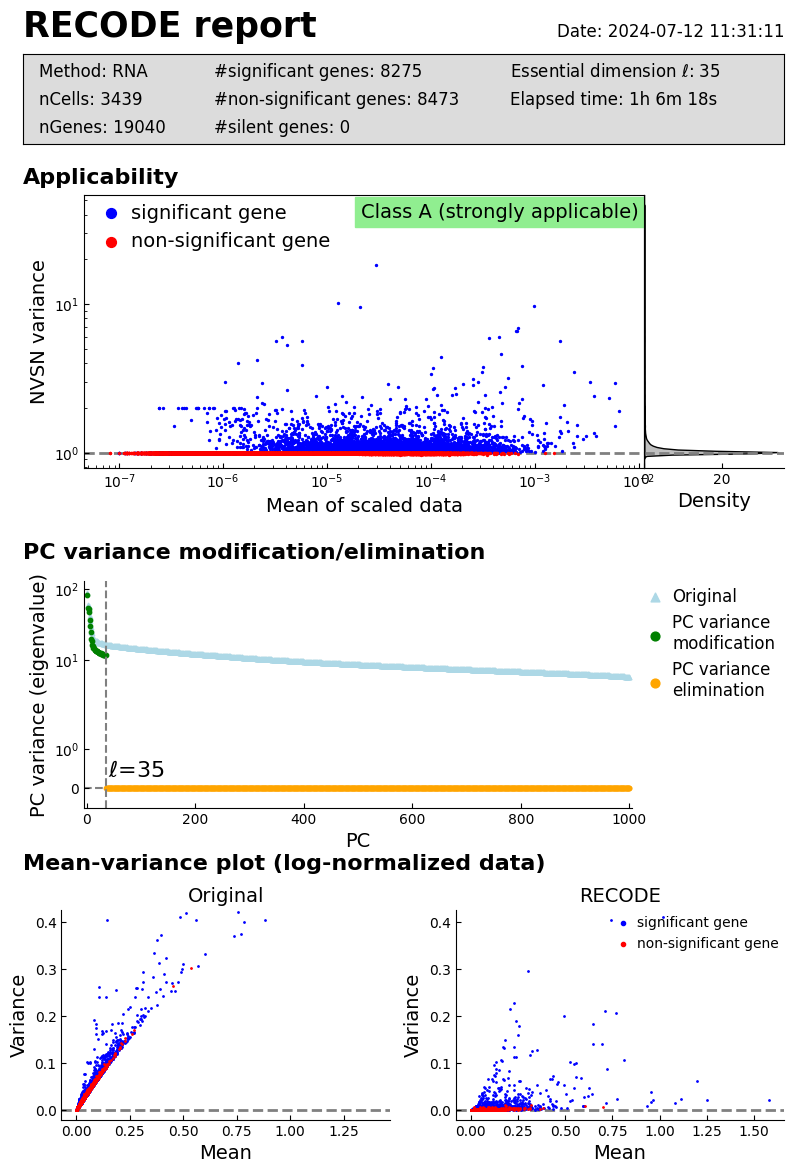
log: {'seq_target': 'RNA', '#significant genes': 8275, '#non-significant genes': 8473, '#silent genes': 0, 'ell': 35, 'Elapsed time': '1h 6m 18s 501ms', 'solver': 'randomized', '#test_data': 3439}
Performance check
[4]:
recode.report()
Log normalizaation
[5]:
target_sum = np.median(np.sum(adata.layers["RECODE"],axis=1))
adata = recode.lognormalize(adata,target_sum=target_sum)
print(np.median(np.sum(adata.layers["RECODE"],axis=1)))
Normalized data are stored in "RECODE_norm" and "RECODE_log"
687.3703800000001
[6]:
adata.layers["Raw_norm"] = target_sum*adata.layers["Raw"]/np.sum(adata.layers["Raw"],axis=1)[:,np.newaxis]
adata.layers["Raw_log"] = np.log(adata.layers["Raw_norm"]+1)
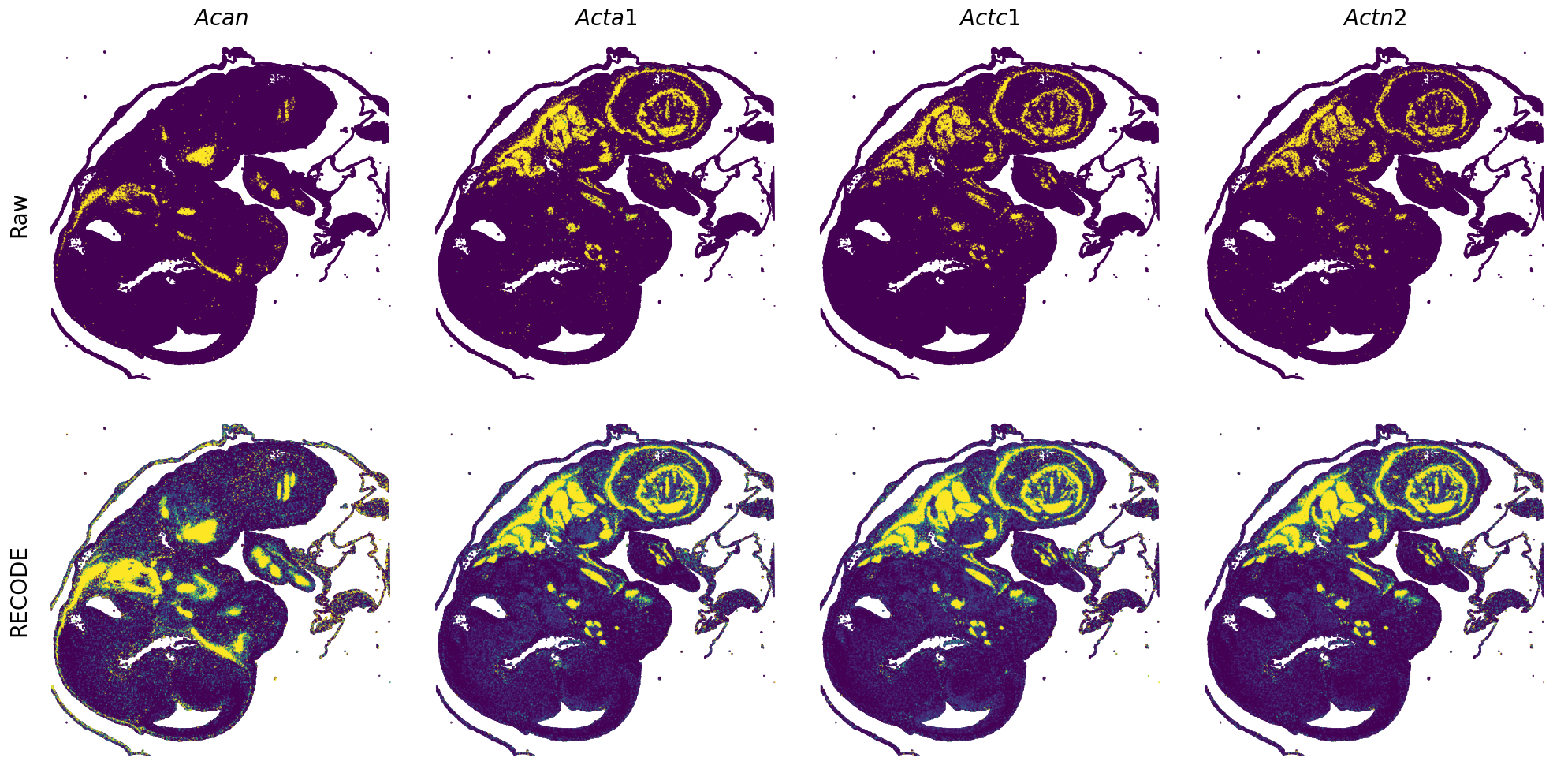
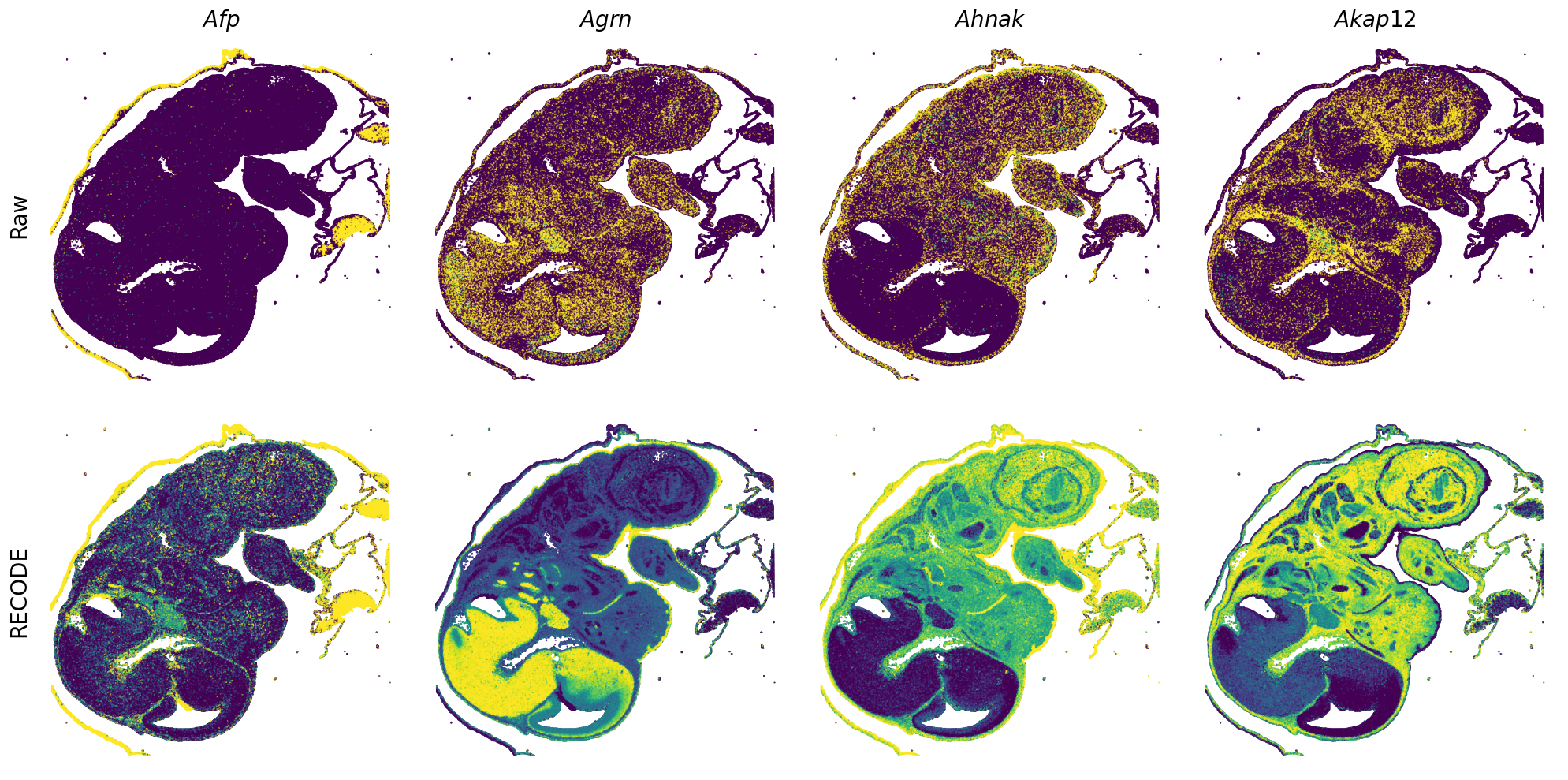
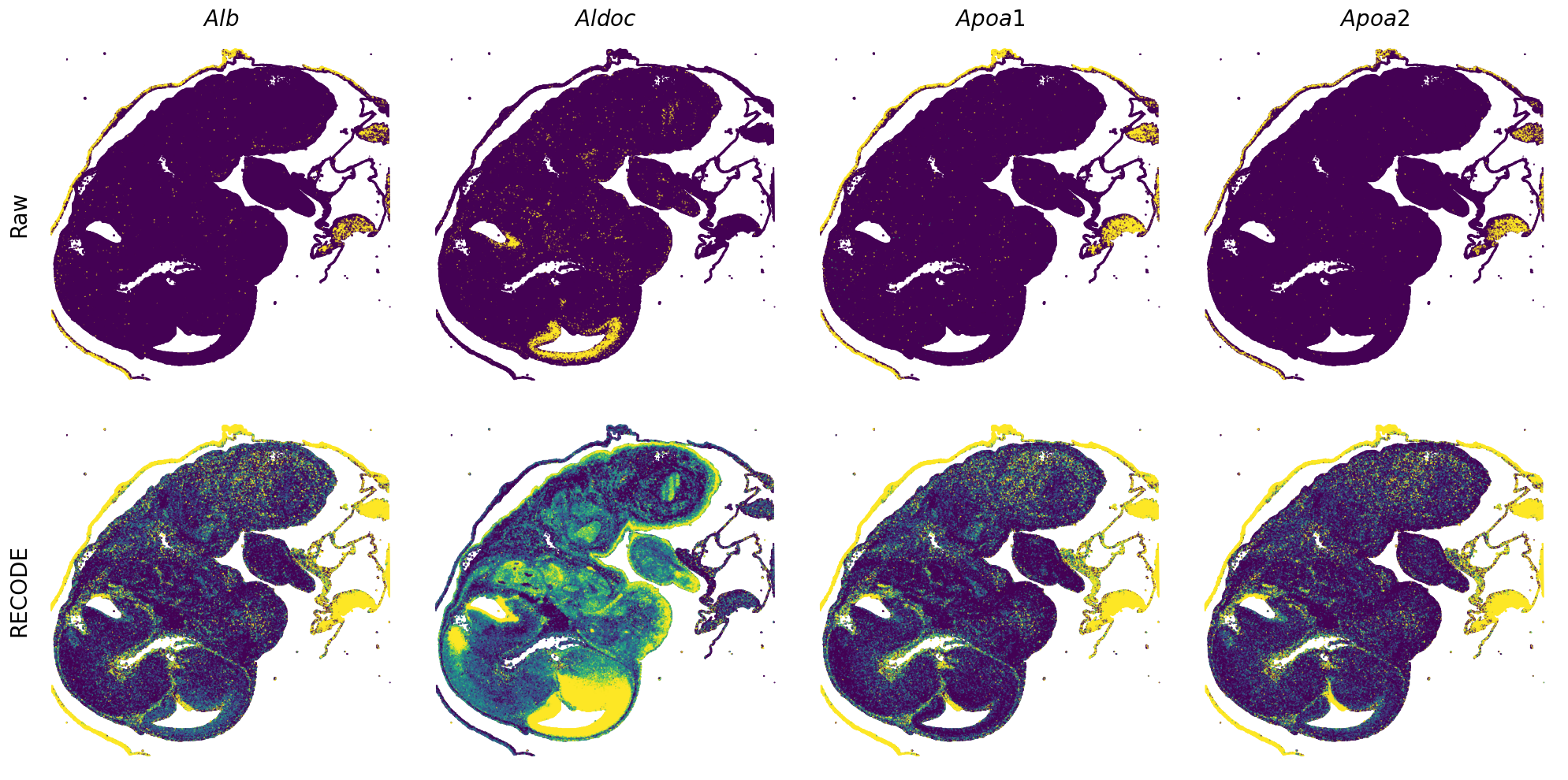
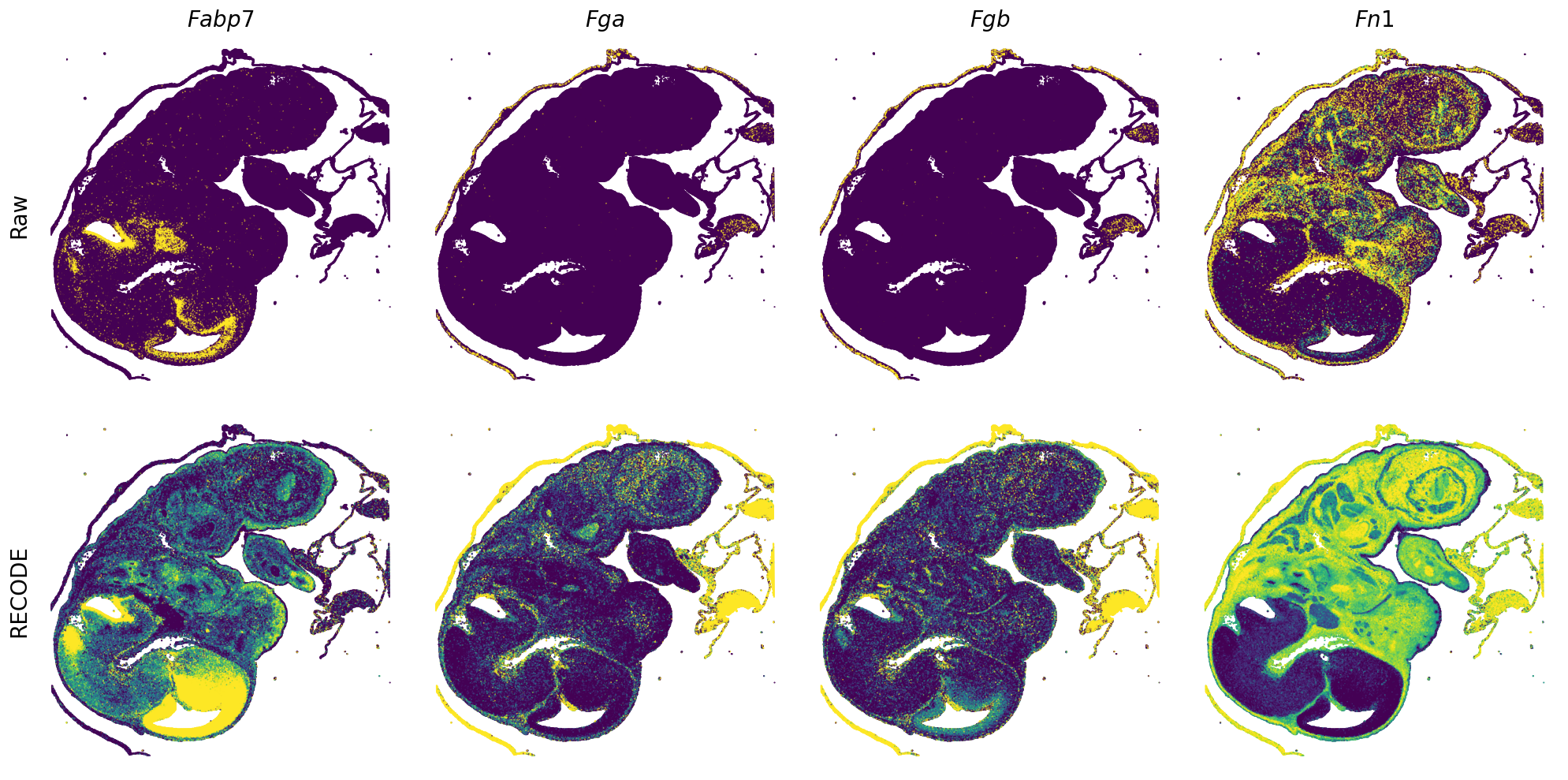
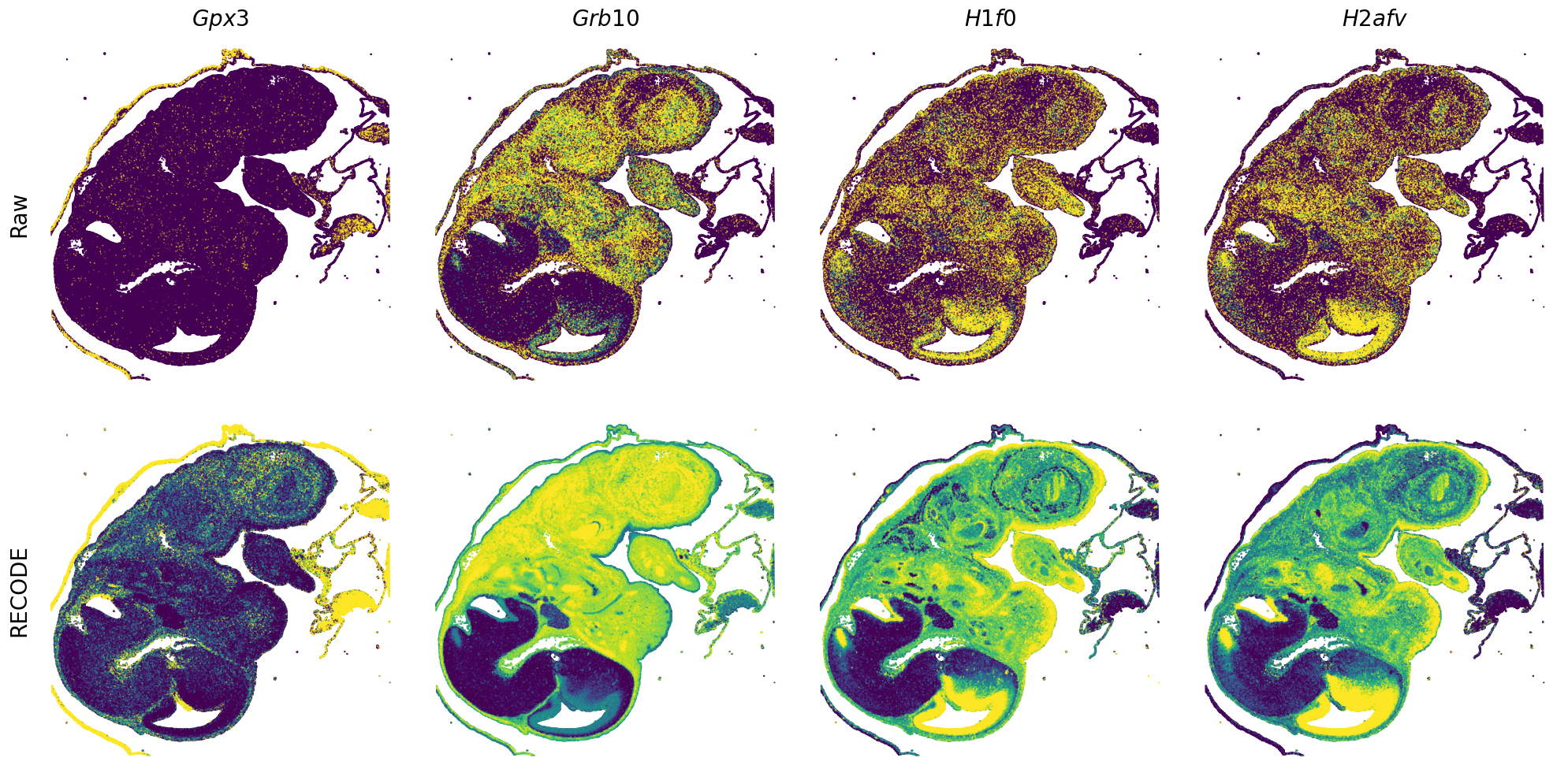
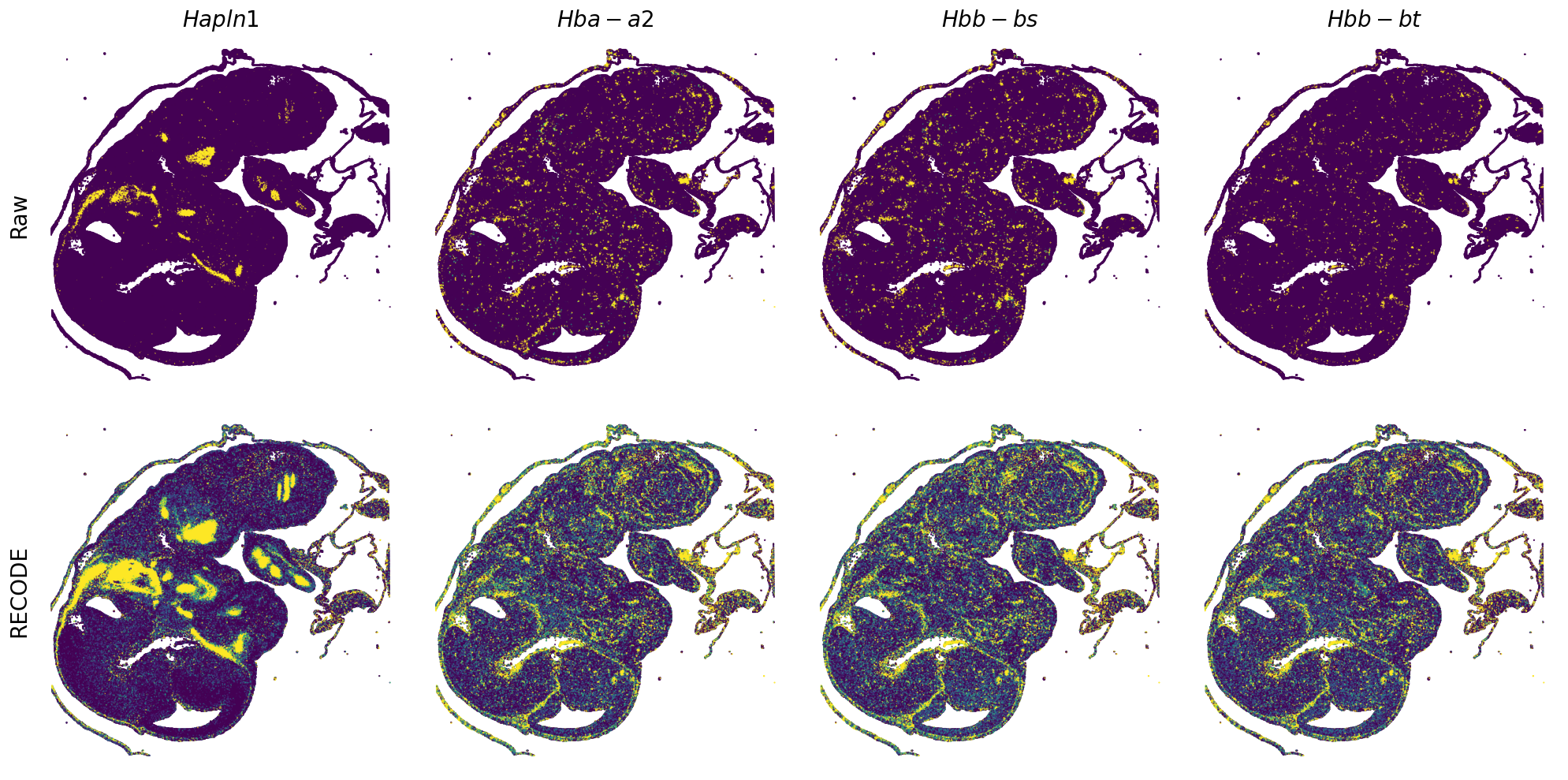
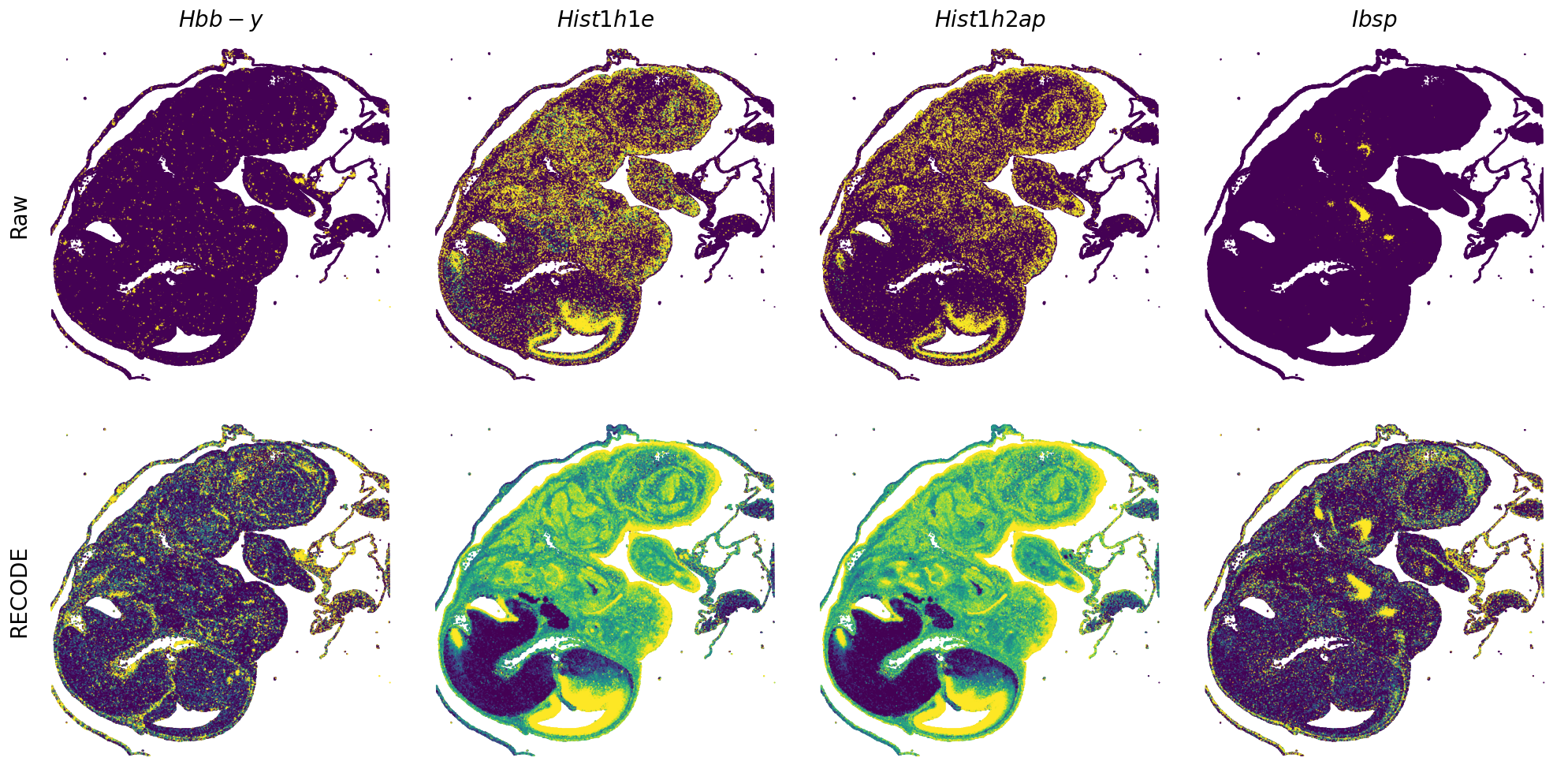
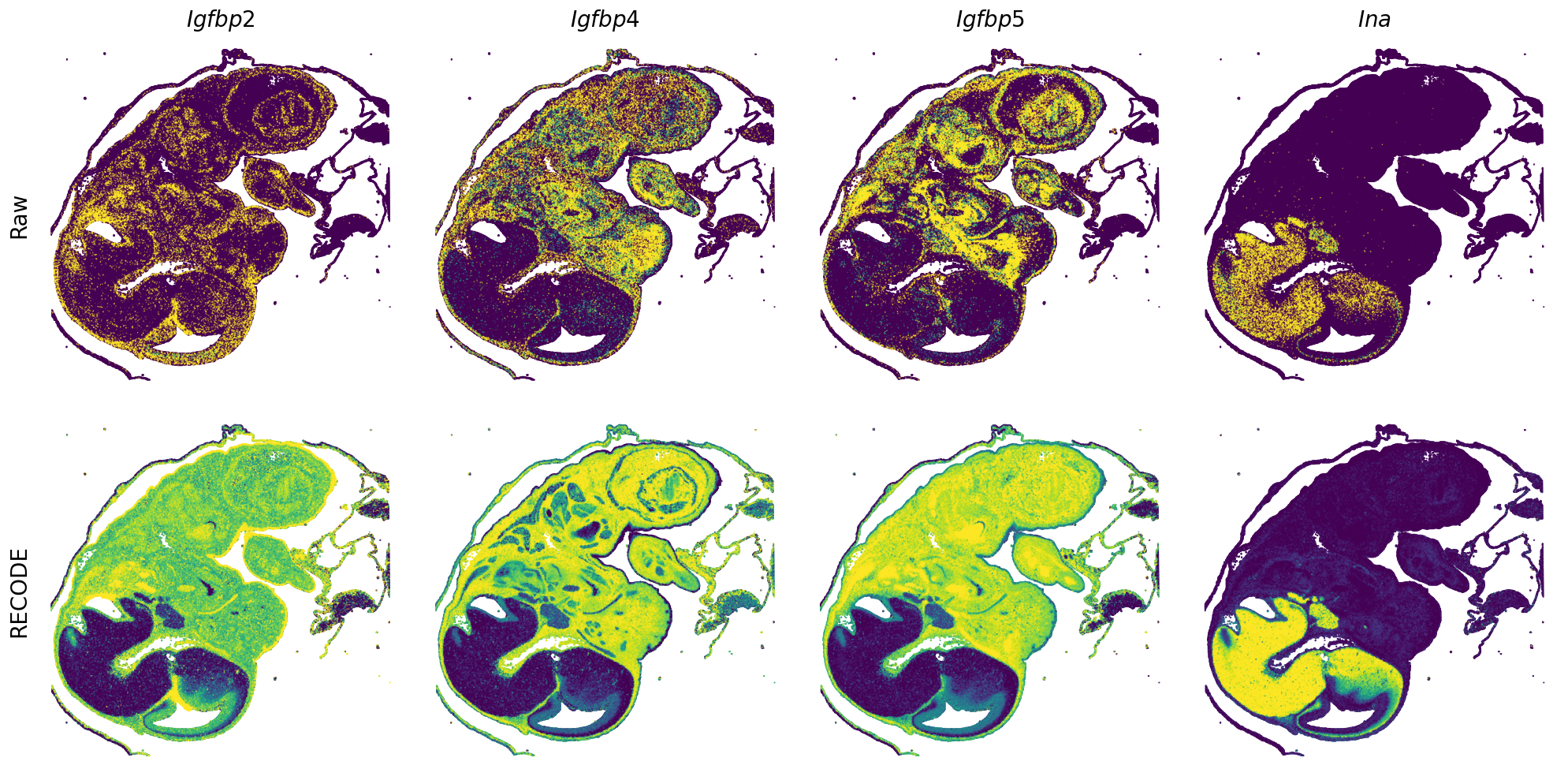
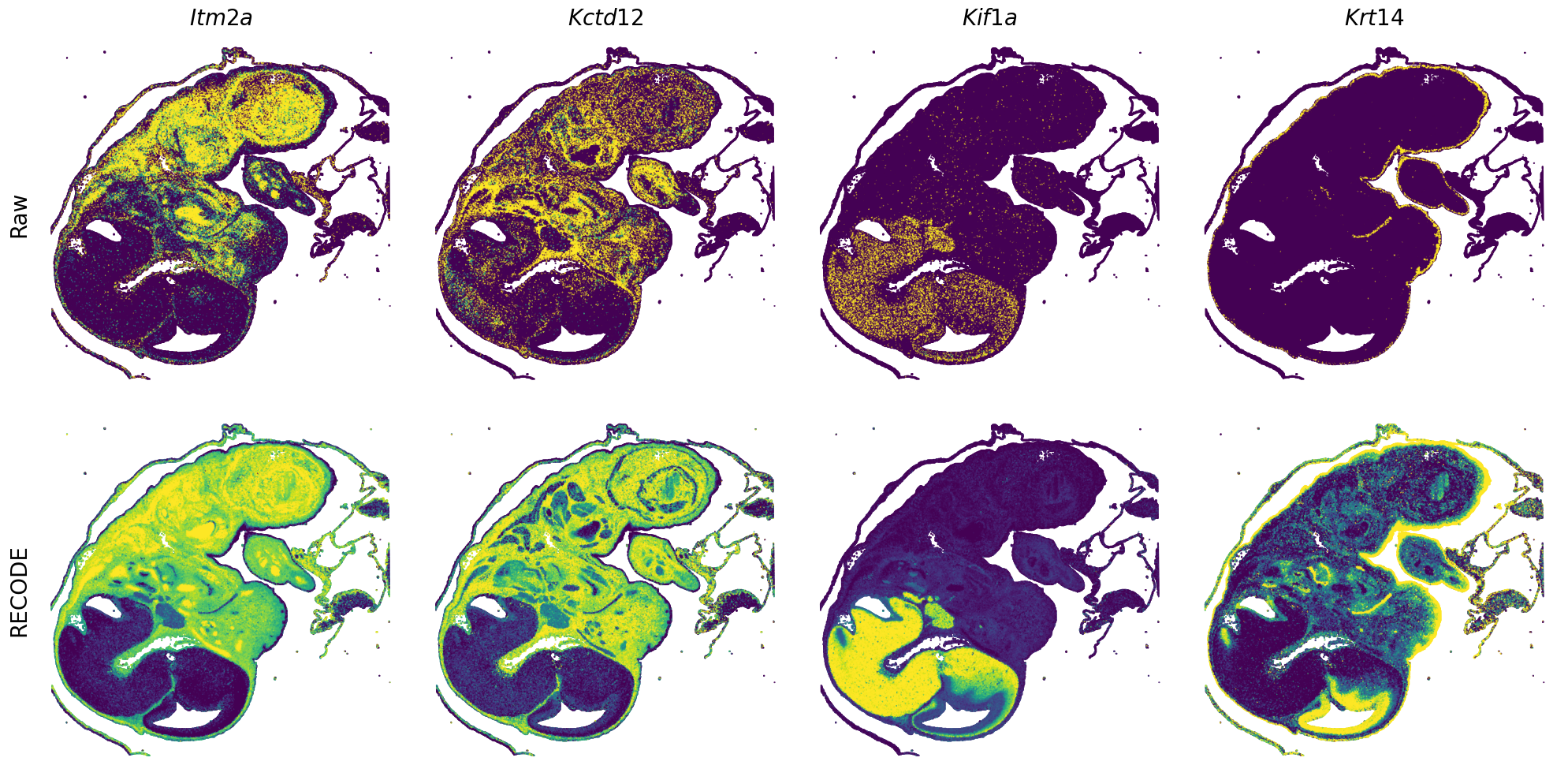
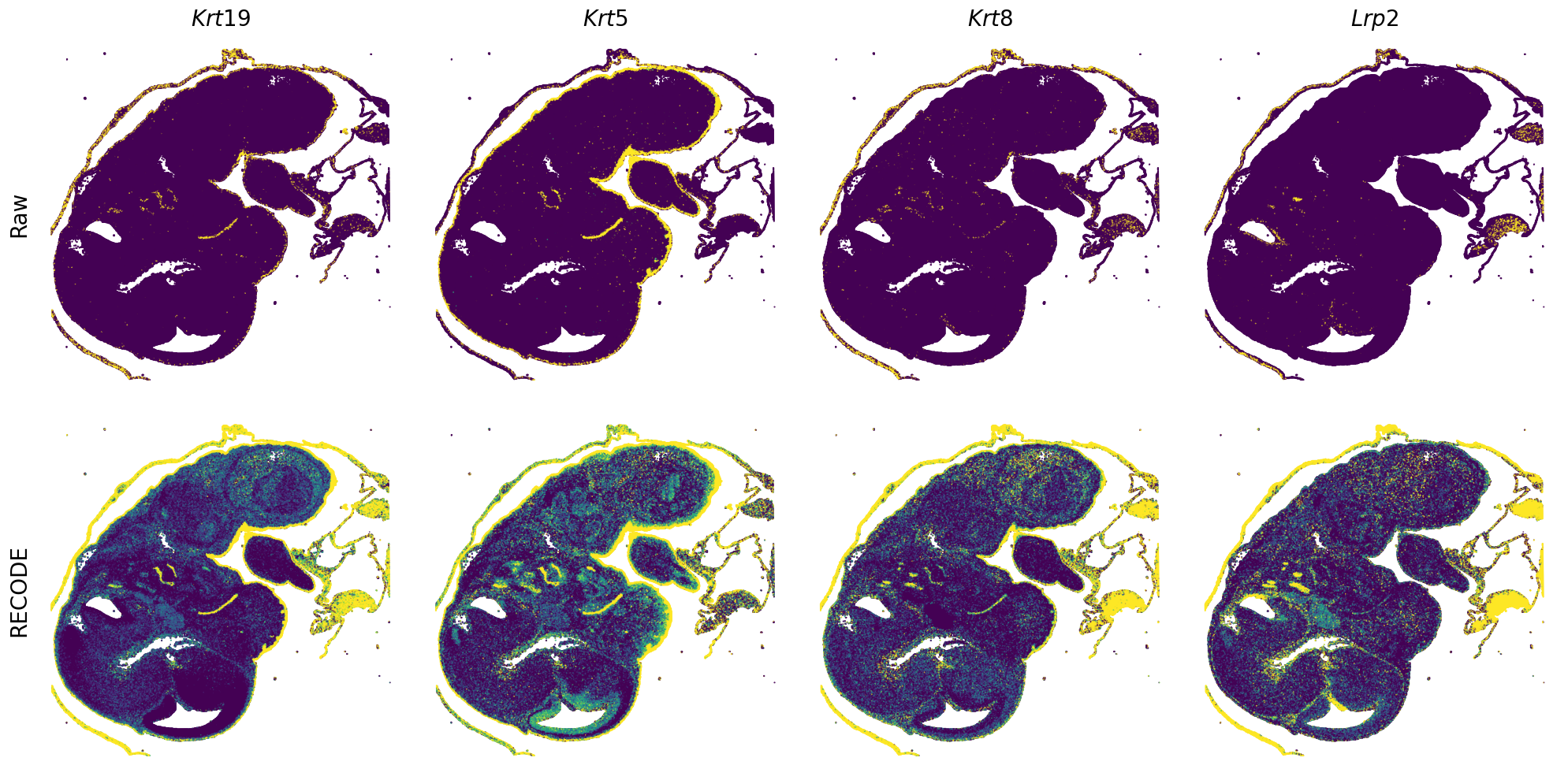
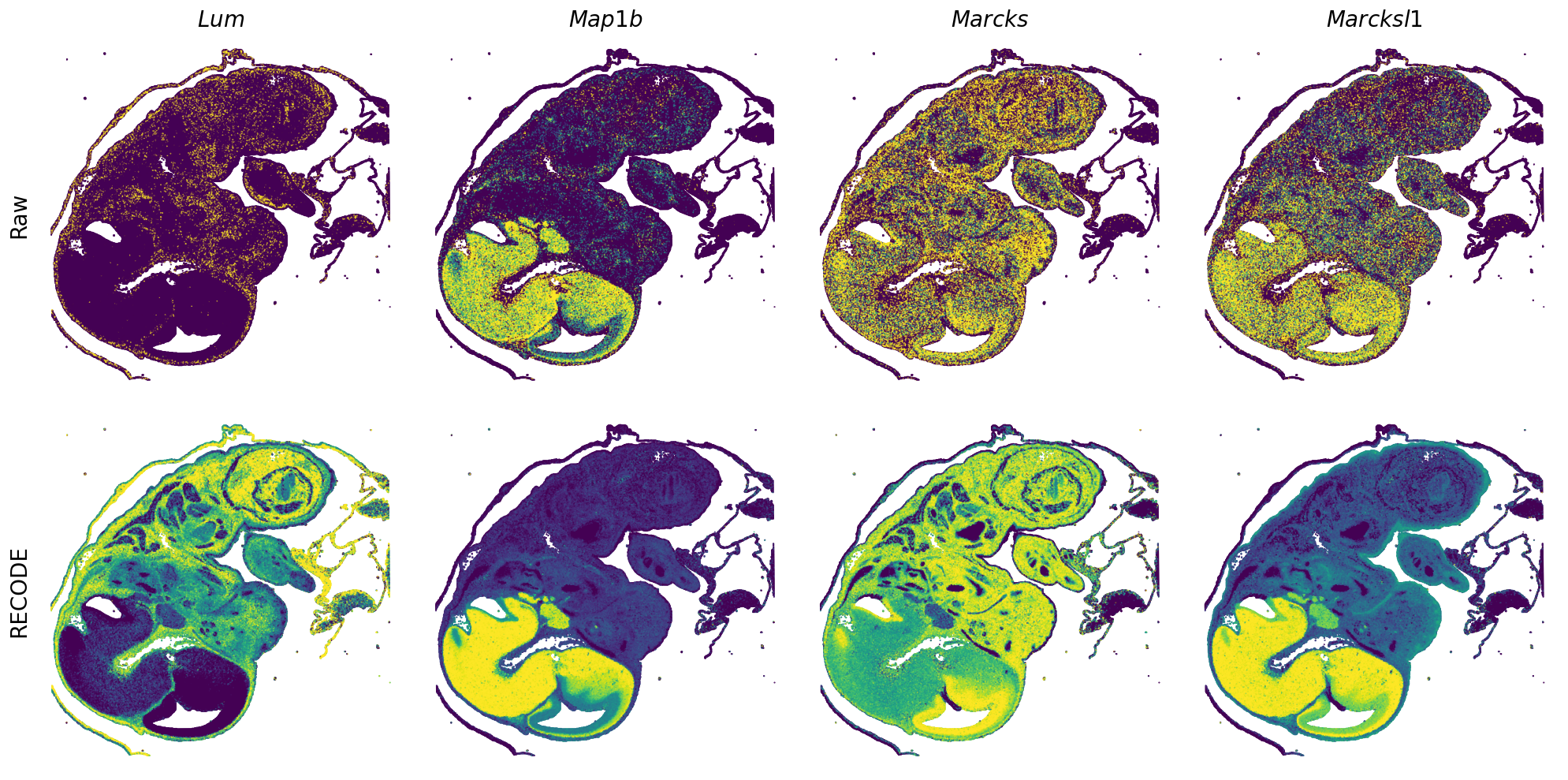
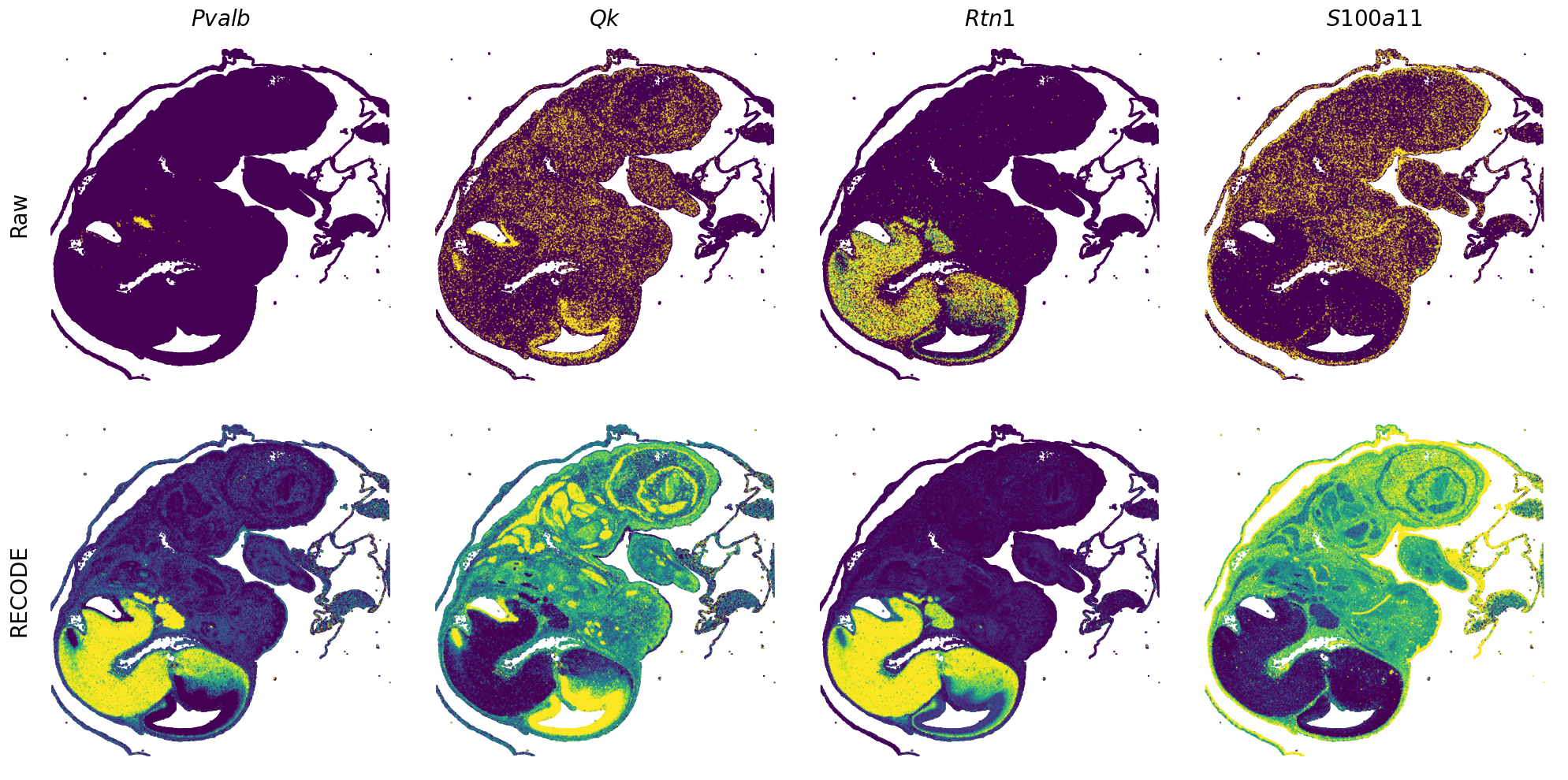
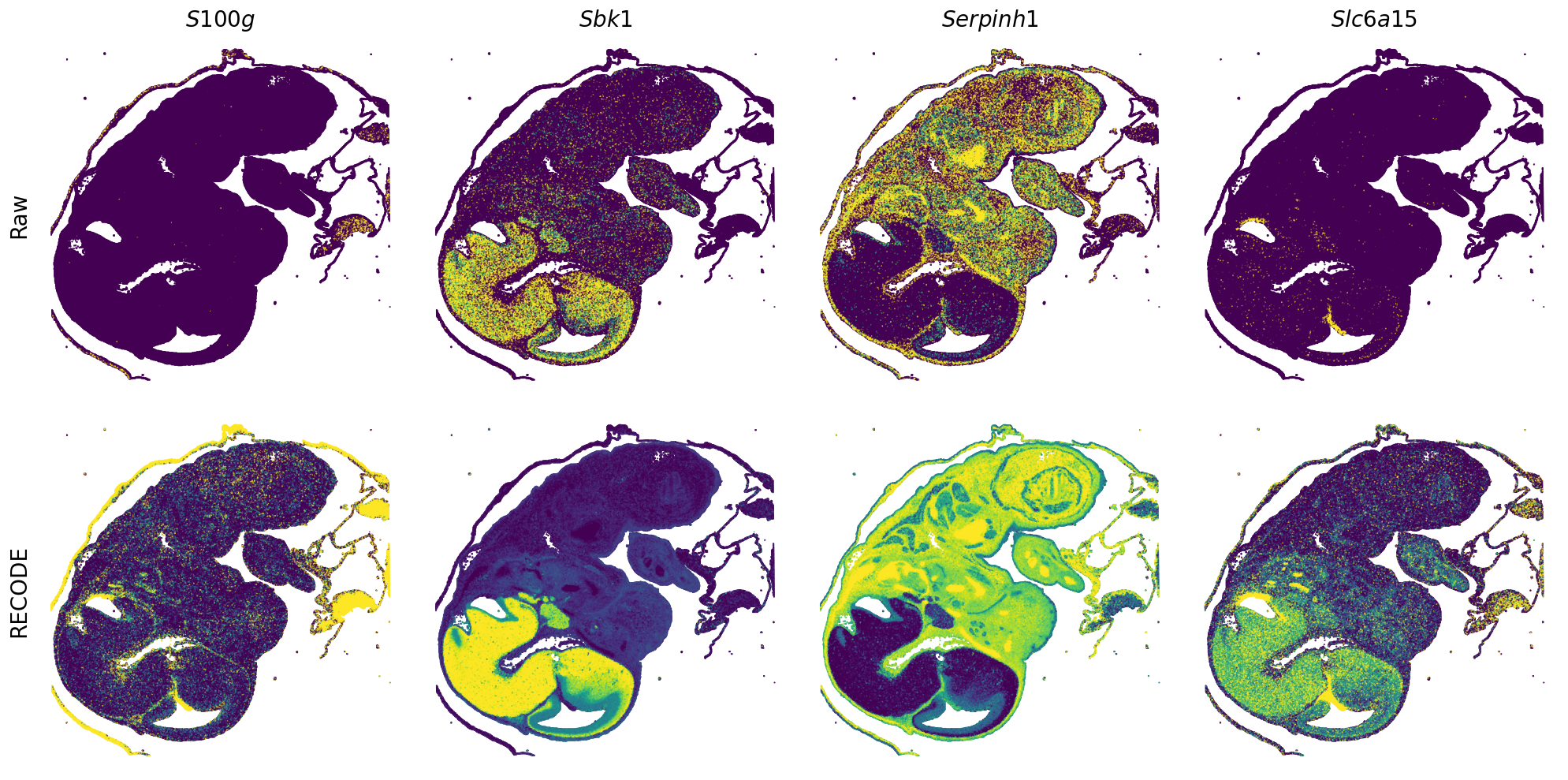
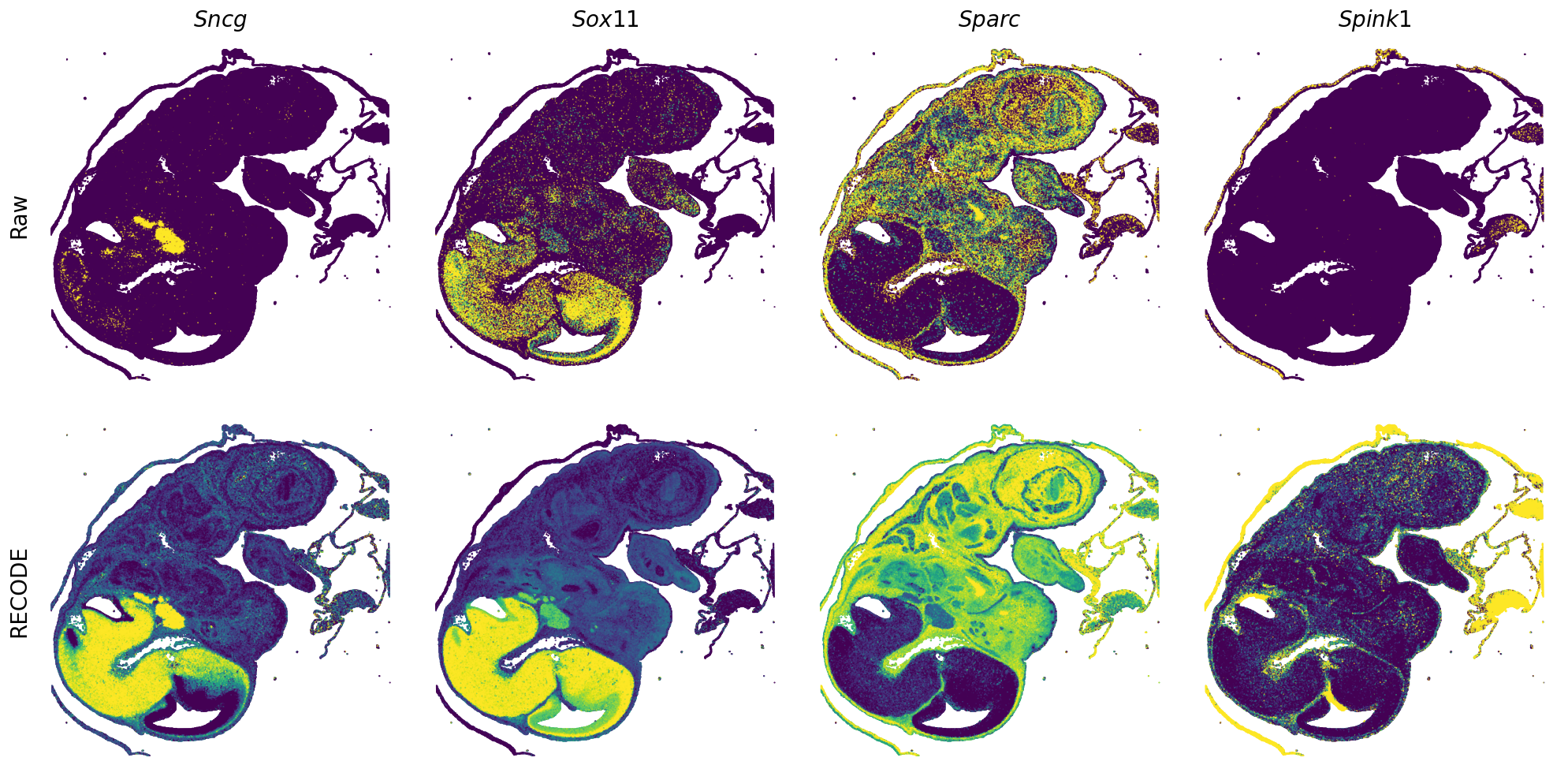
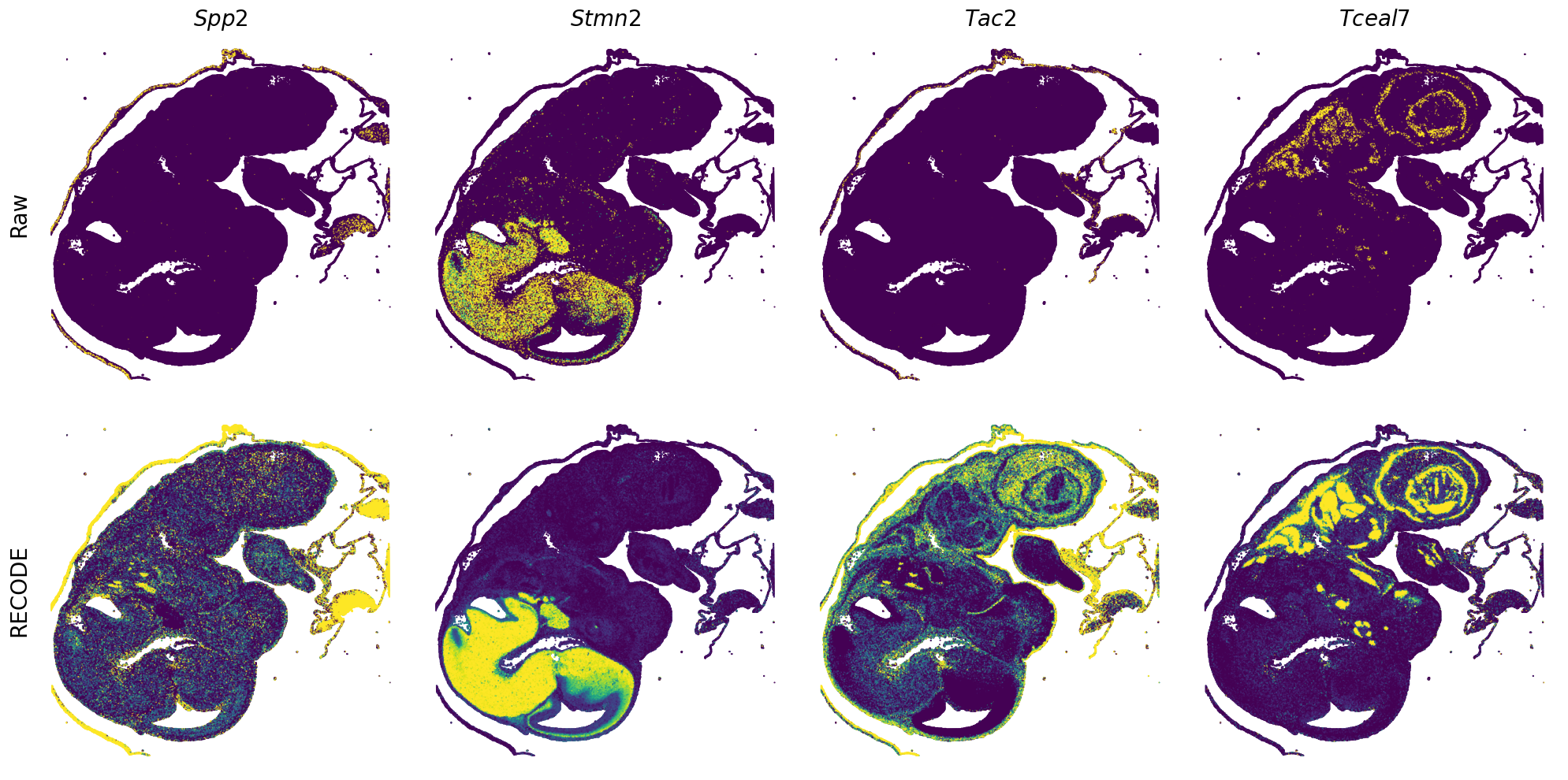
Plot spatial gene expression
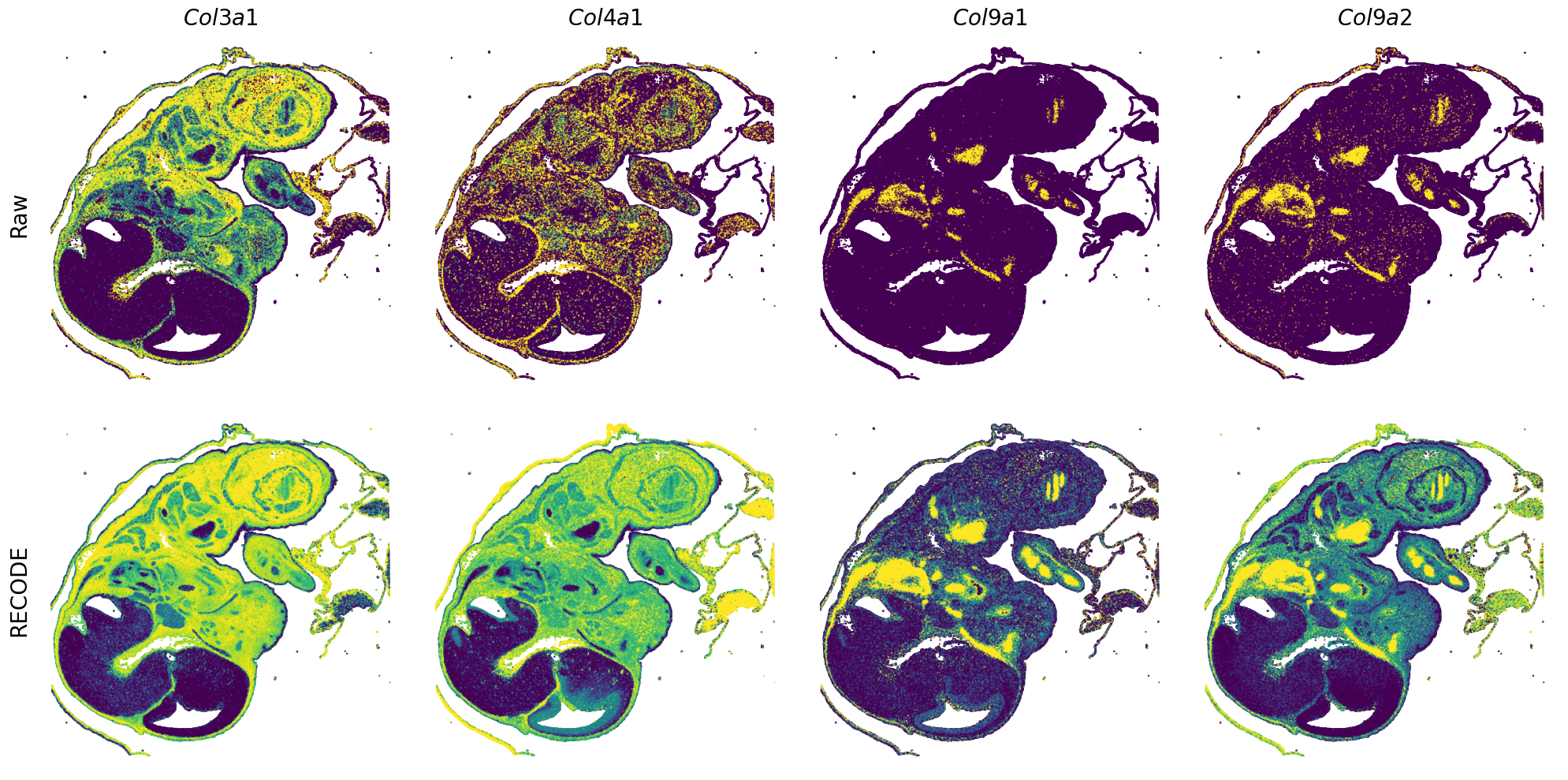
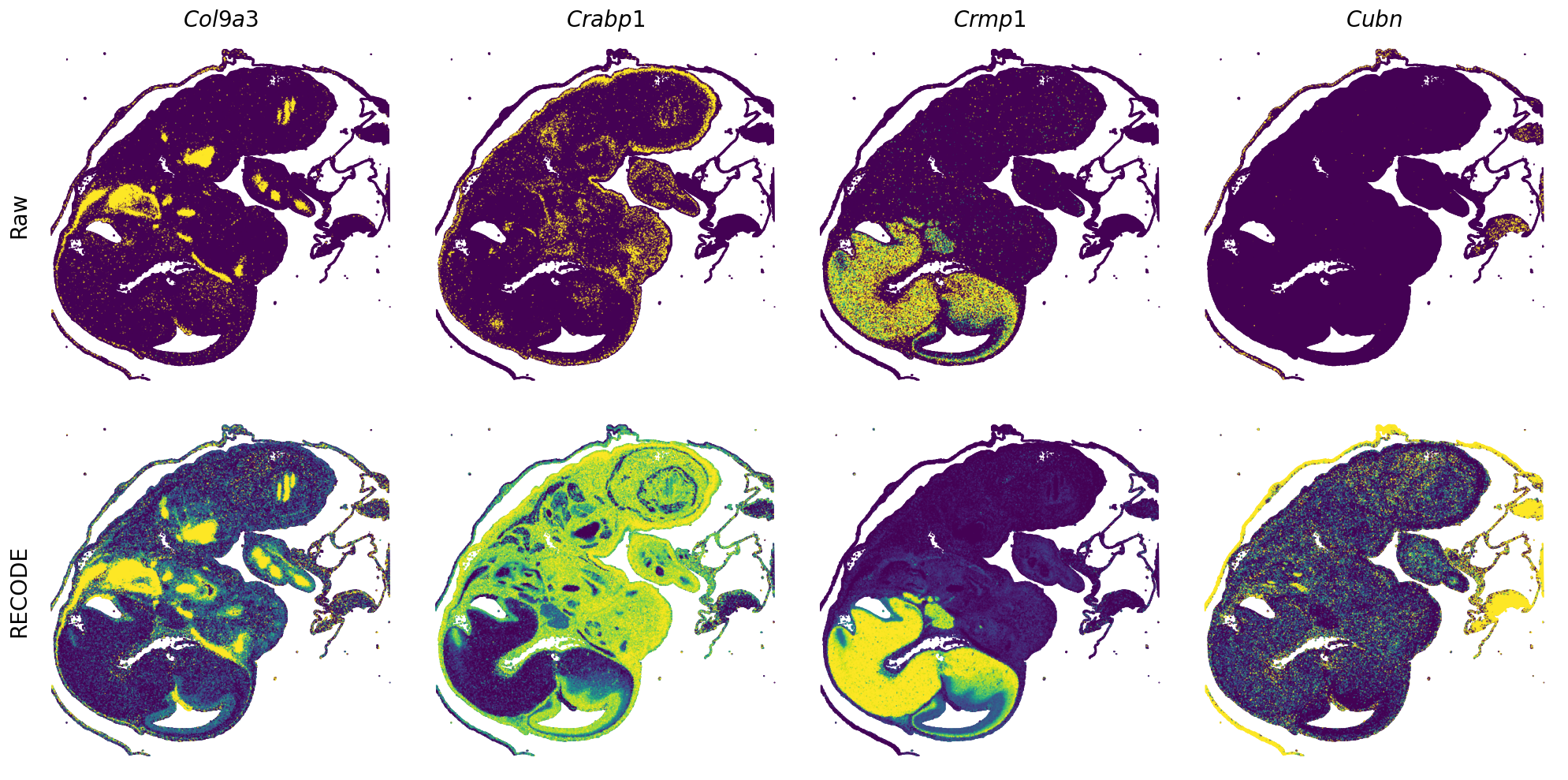
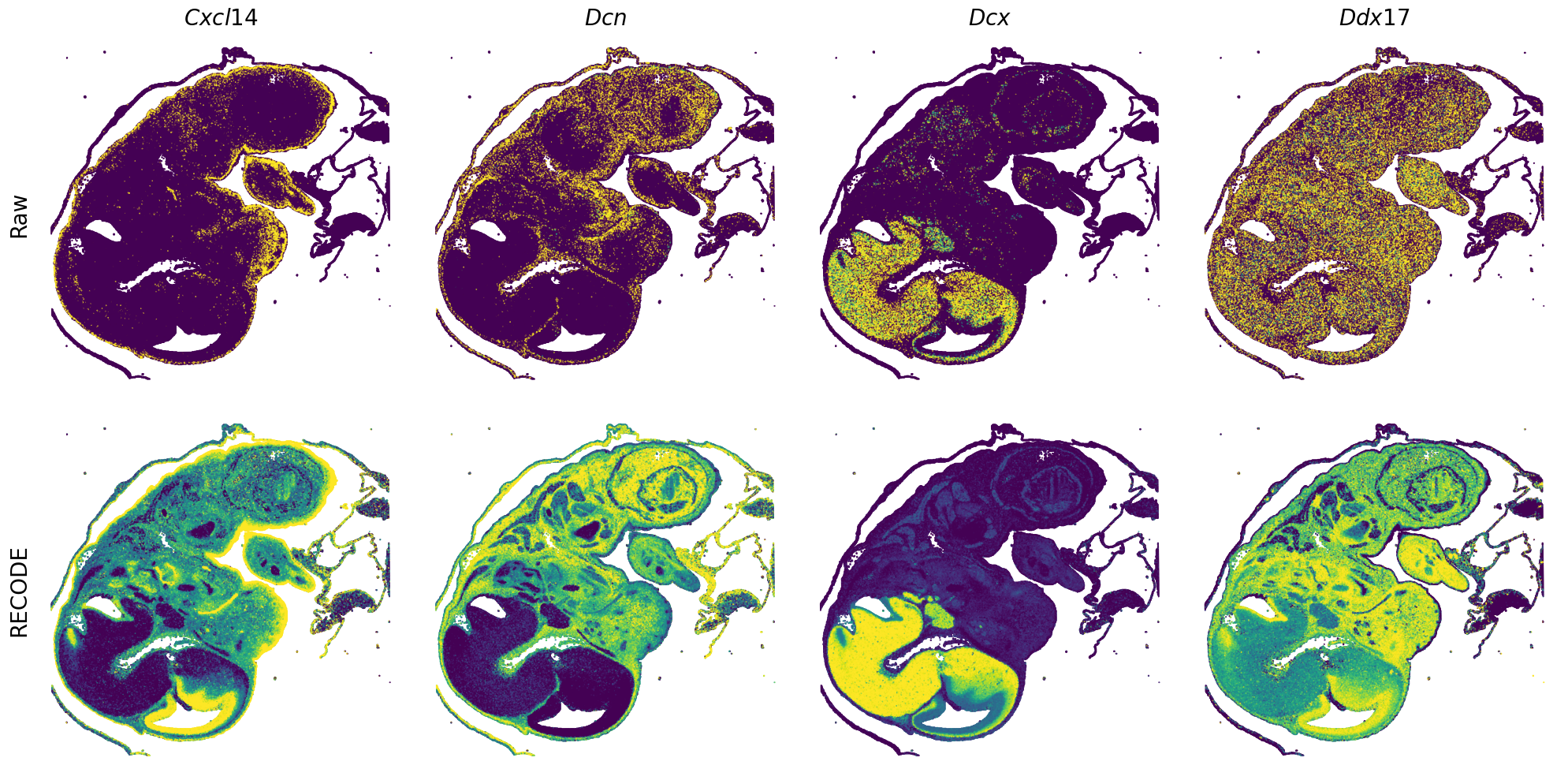
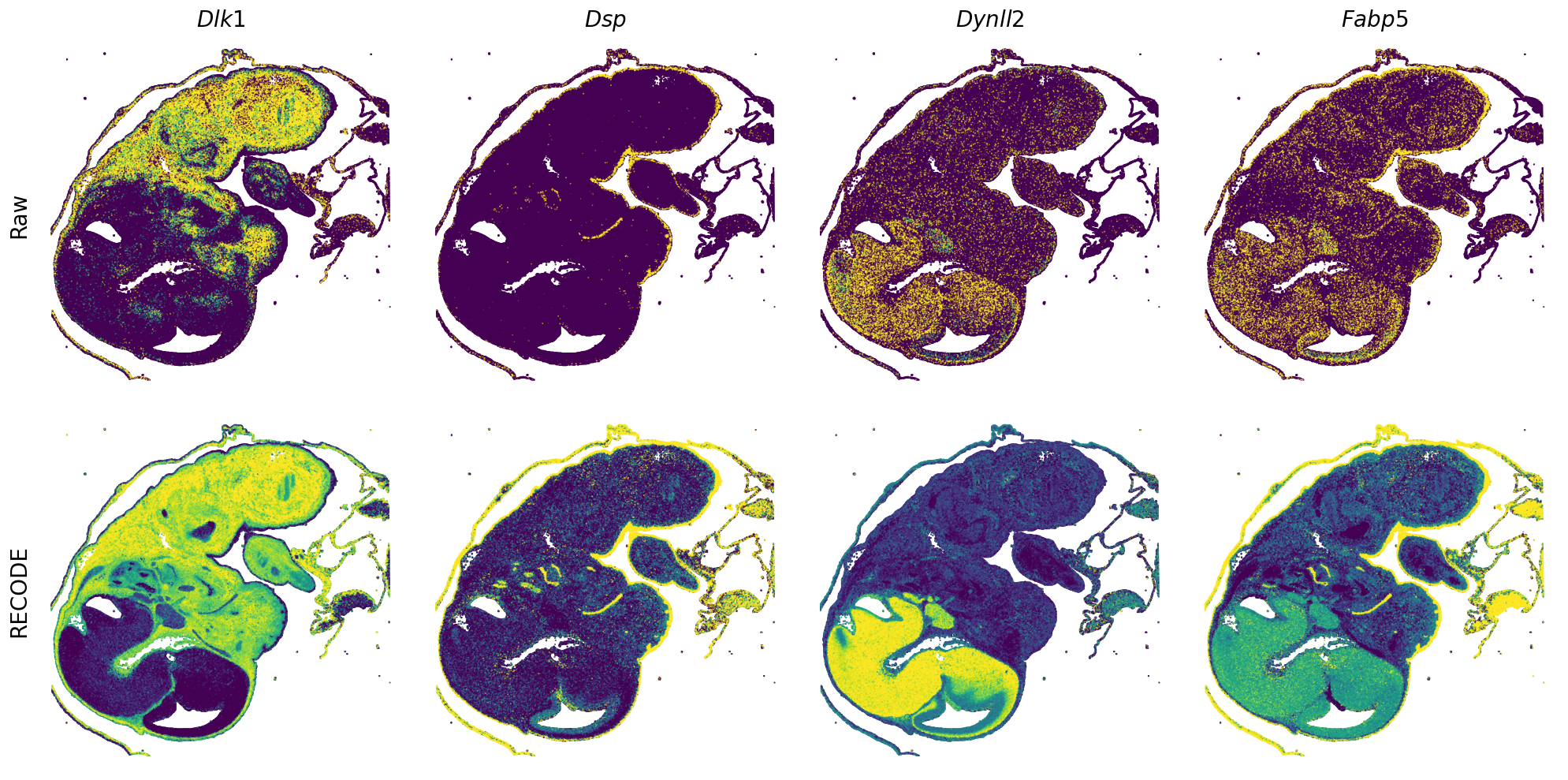
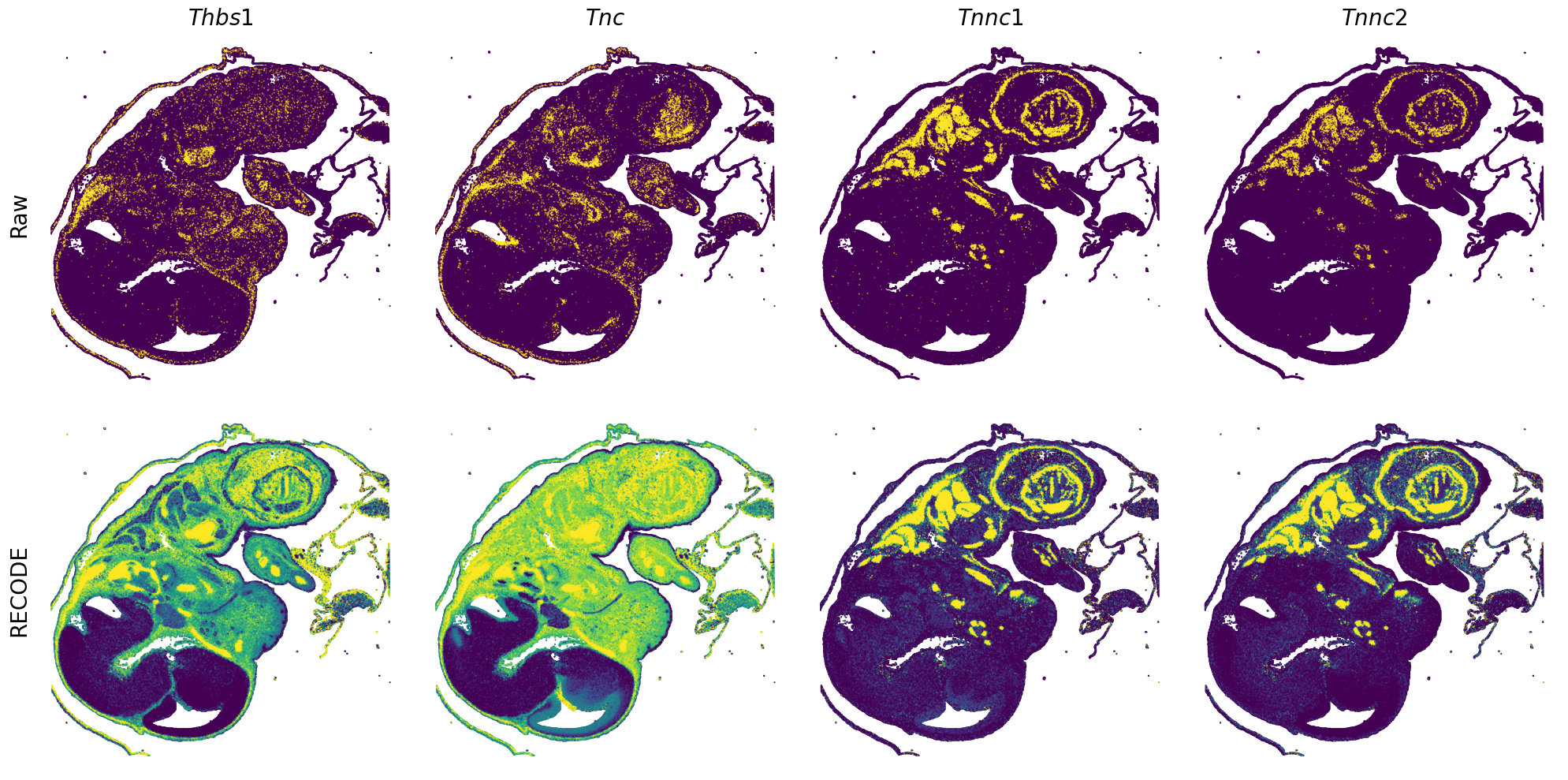
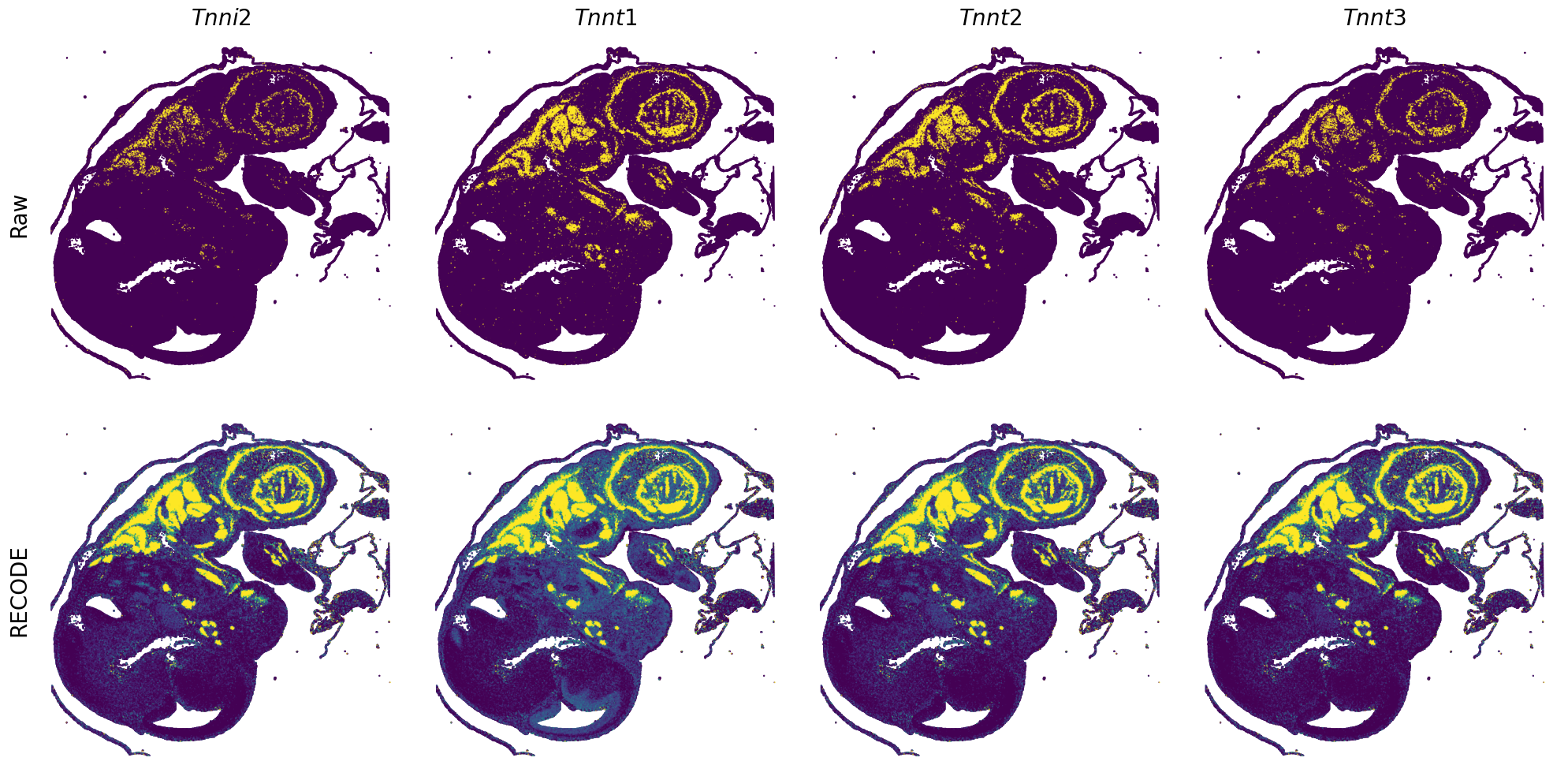
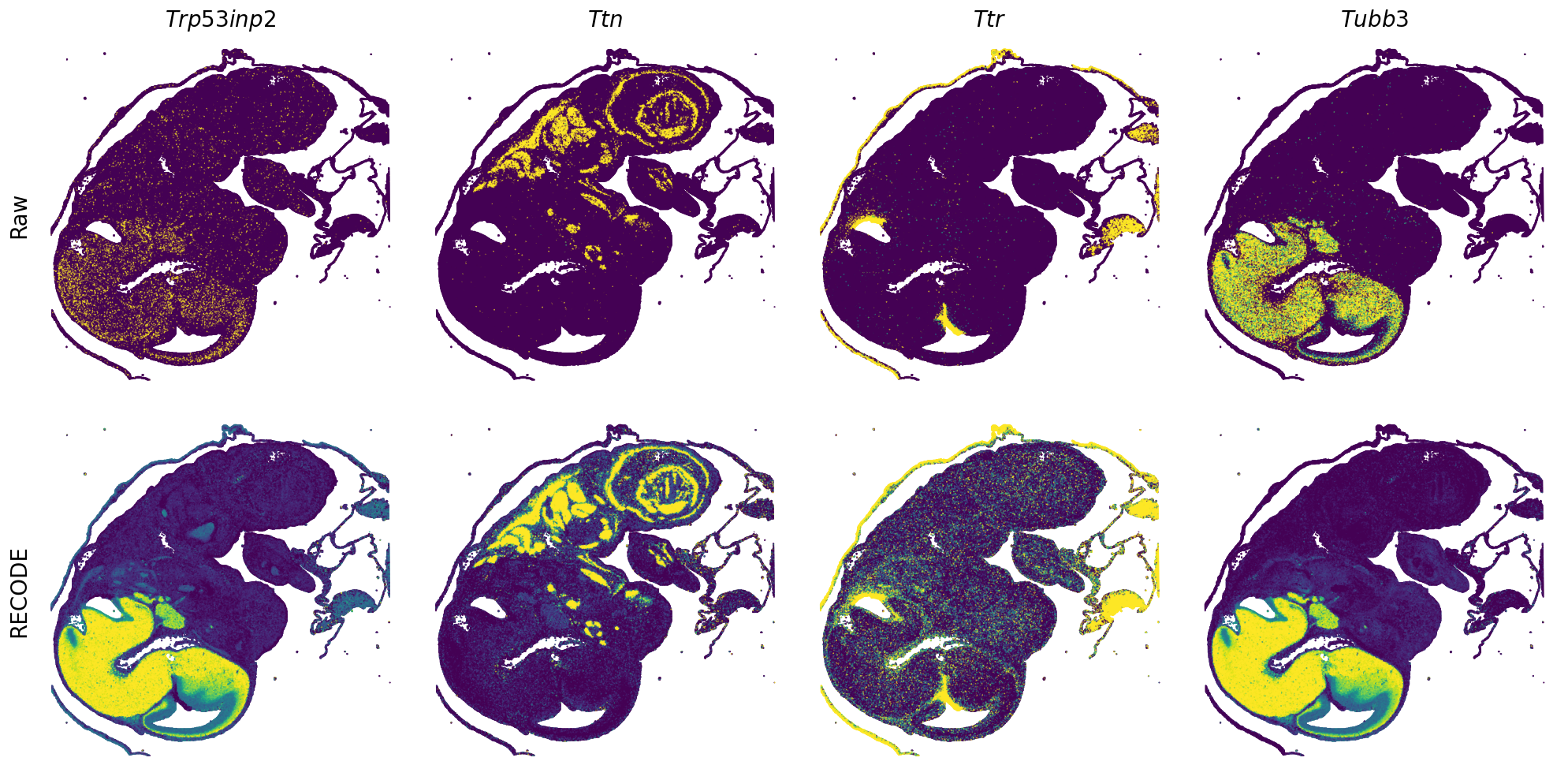
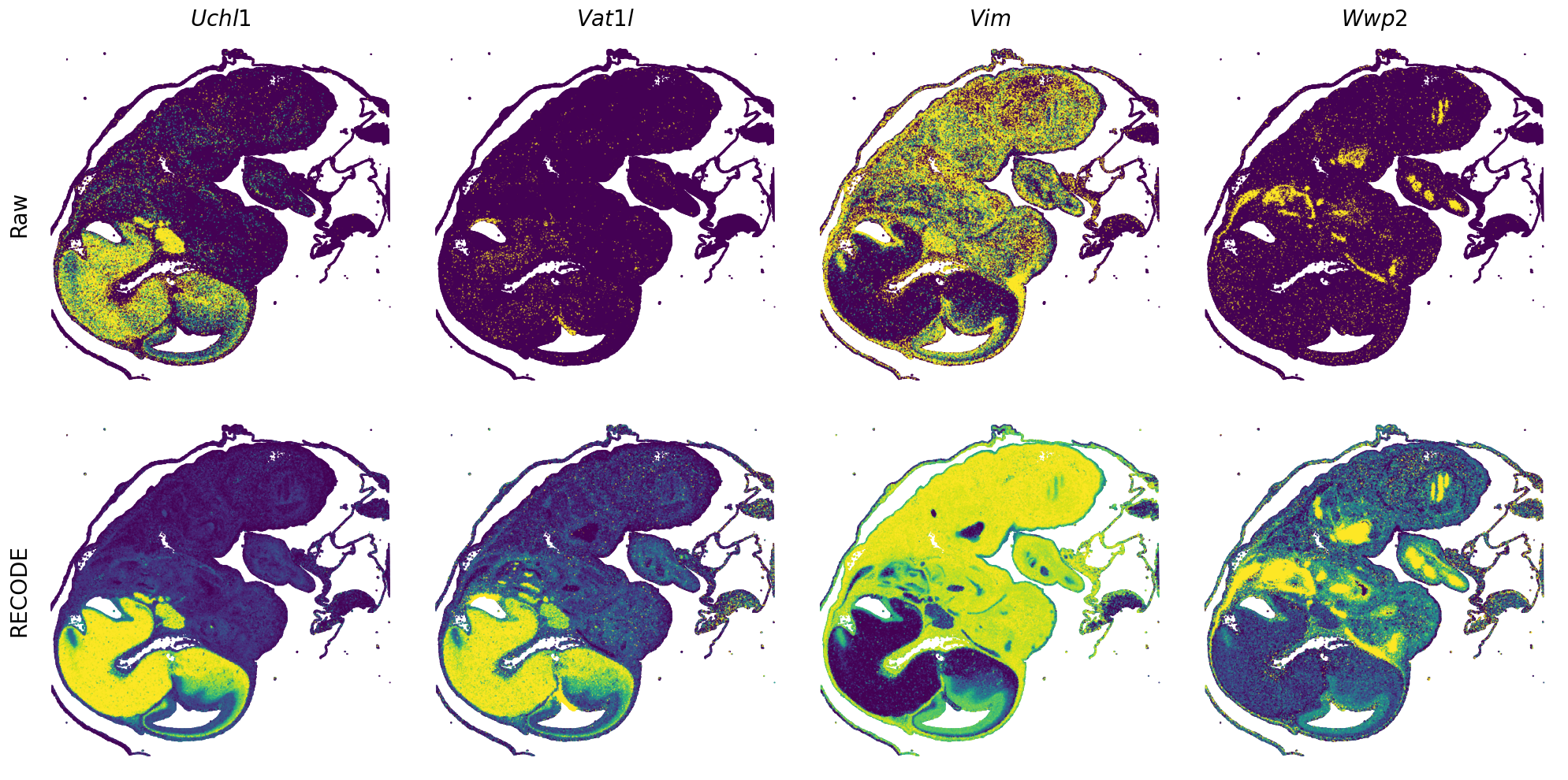
[7]:
def spatial_gex(
genes,
sp_x = adata.obs["pxl_col_in_fullres"],
sp_y = -adata.obs["pxl_row_in_fullres"],
psize = 0.1,
figsize=(5,5),
dpi=100,
percentiles = [10,90],
fs_title = 20,
fs_label = 20,
):
fig,ax = plt.subplots(2,len(genes),figsize=(figsize[0]*len(genes),figsize[1]*2),tight_layout=True)
for i in range(len(genes)):
idx_gene = adata.var.index == genes[i]
if sum(idx_gene) == 0: continue
exp = adata.layers["RECODE_log"][:,idx_gene]
vmin,vmax = np.percentile(exp,percentiles[0]),np.percentile(exp,percentiles[1])
ax_ = ax[1,i]
ax_.scatter(sp_x, sp_y,c=exp,s=psize,marker="H",vmin=vmin,vmax=vmax)
if i== 0:
ax_.set_ylabel("RECODE",fontsize=fs_label)
ax_.tick_params(bottom=False, left=False, right=False, top=False,
labelbottom=False, labelleft=False, labelright=False, labeltop=False)
[ax_.spines[c_].set_visible(False) for c_ in ['right','top','bottom','left']]
else:
ax_.axis('off')
exp = adata.layers["Raw_log"][:,idx_gene]
ax_ = ax[0,i]
ax_.scatter(sp_x, sp_y,c=exp,s=psize,marker="H",vmin=vmin,vmax=vmax)
ax_.set_title("$\it{%s}$" % genes[i],fontsize=fs_title)
if i== 0:
ax_.set_ylabel("Raw",fontsize=fs_label)
ax_.tick_params(bottom=False, left=False, right=False, top=False,
labelbottom=False, labelleft=False, labelright=False, labeltop=False)
[ax_.spines[c_].set_visible(False) for c_ in ['right','top','bottom','left']]
else:
ax_.axis('off')
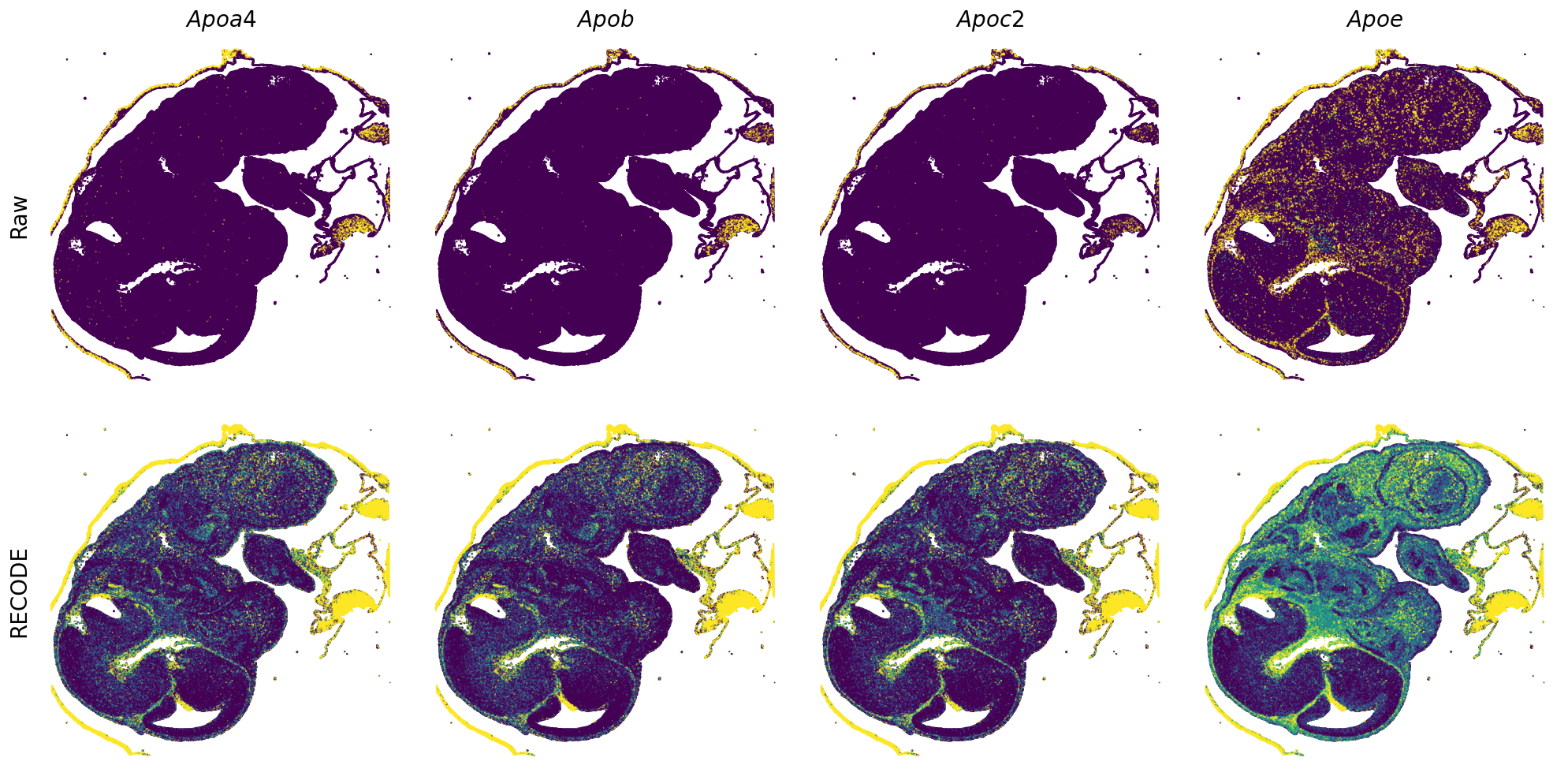
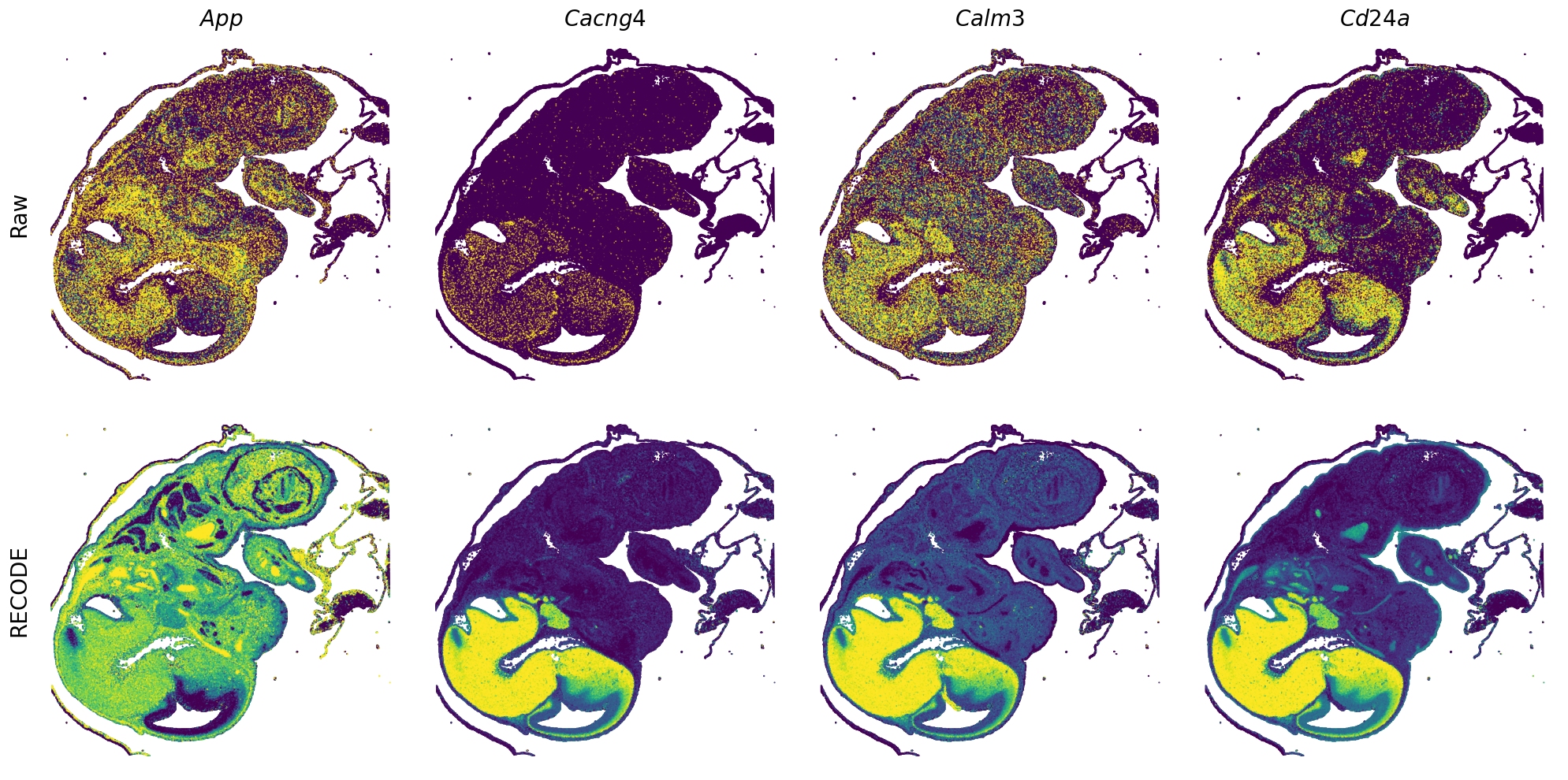
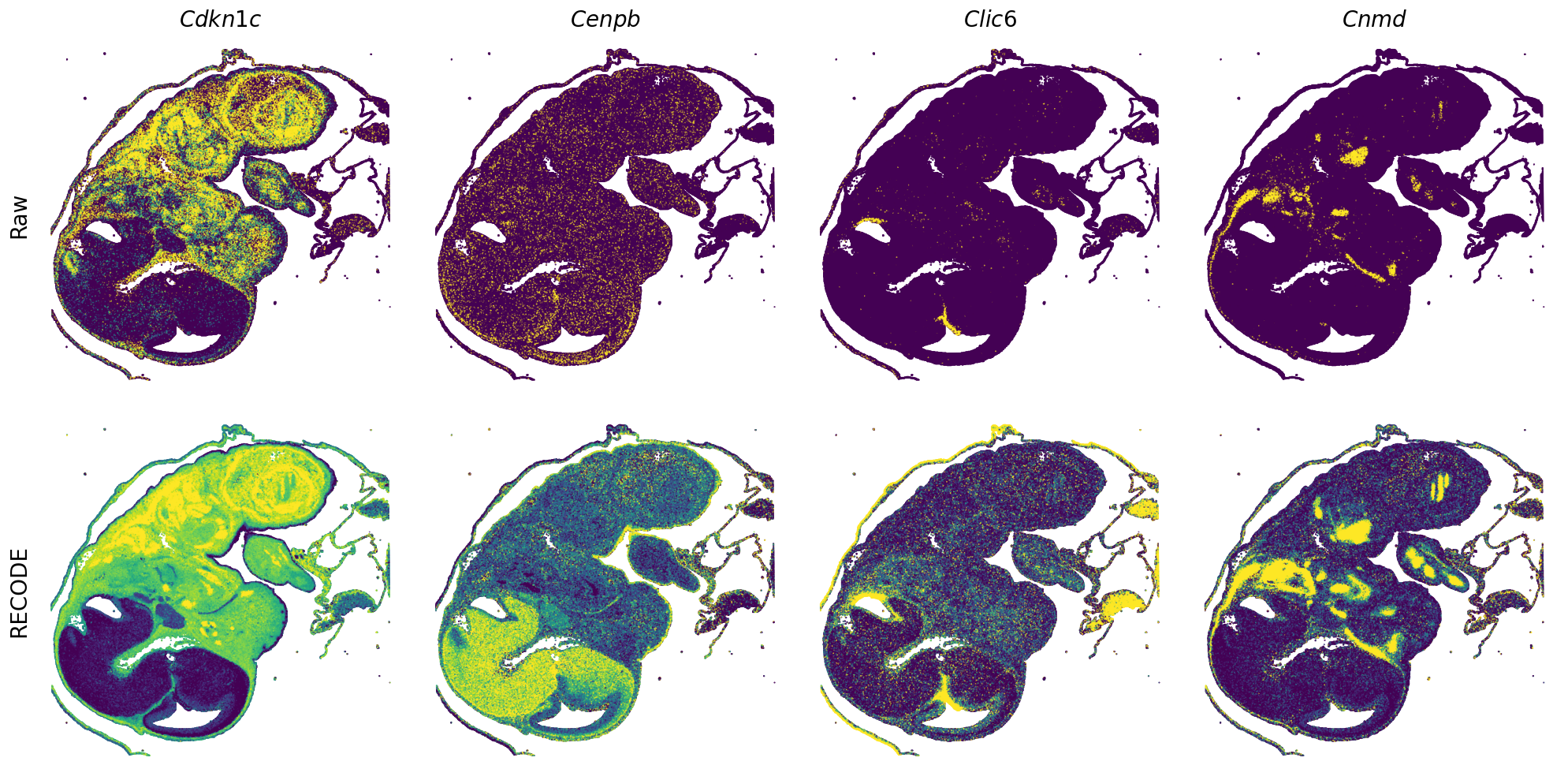
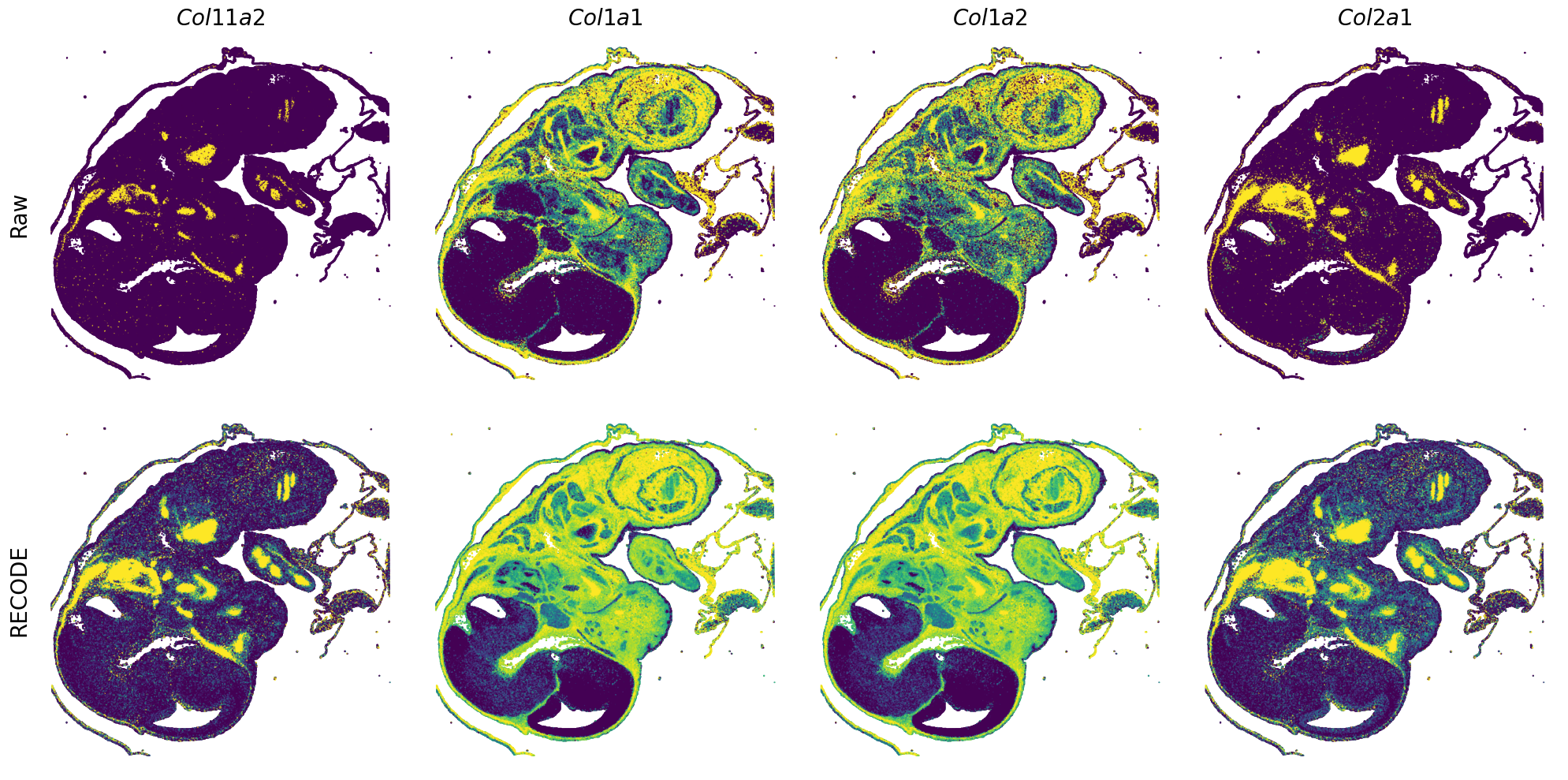
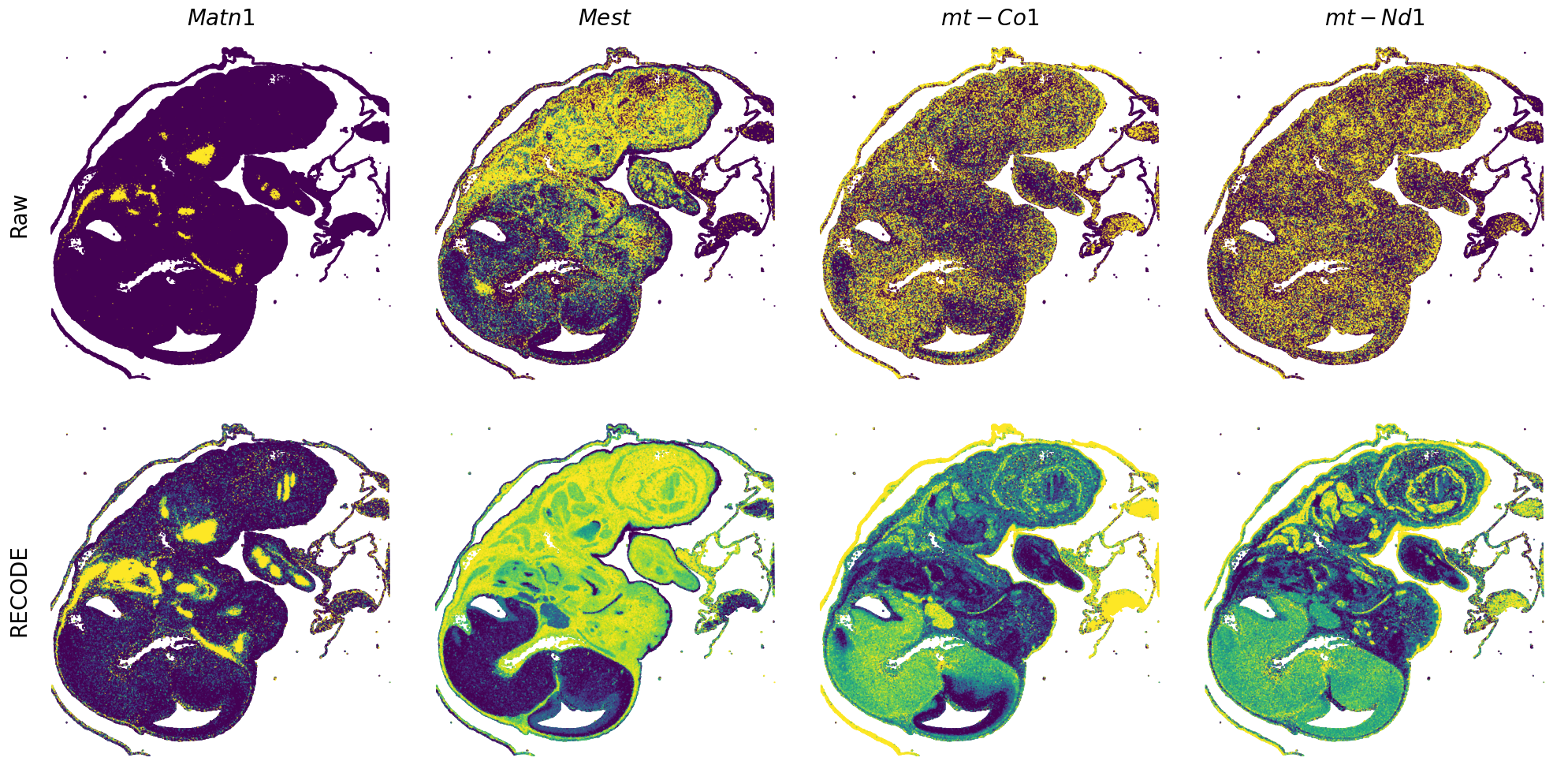
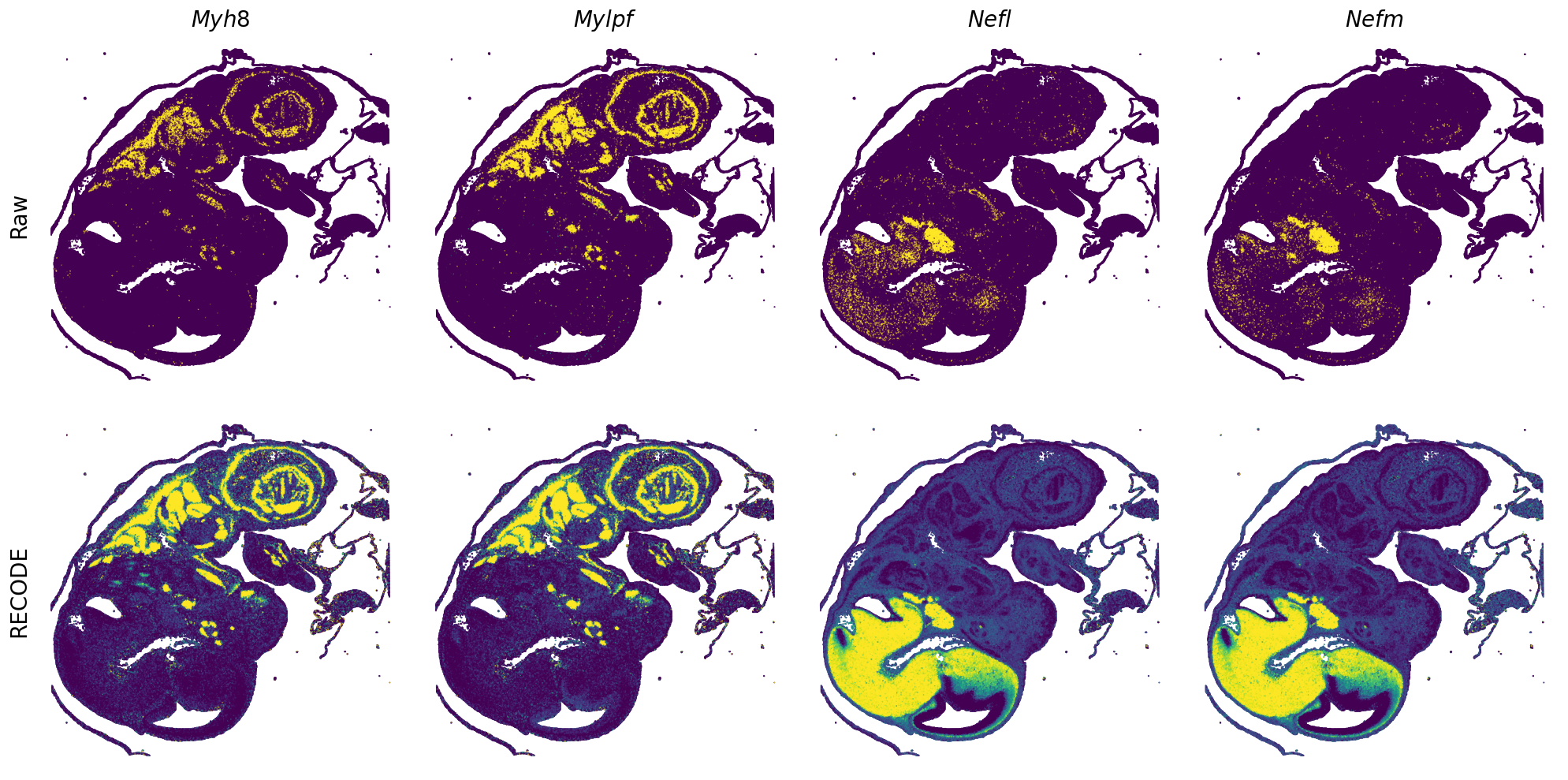
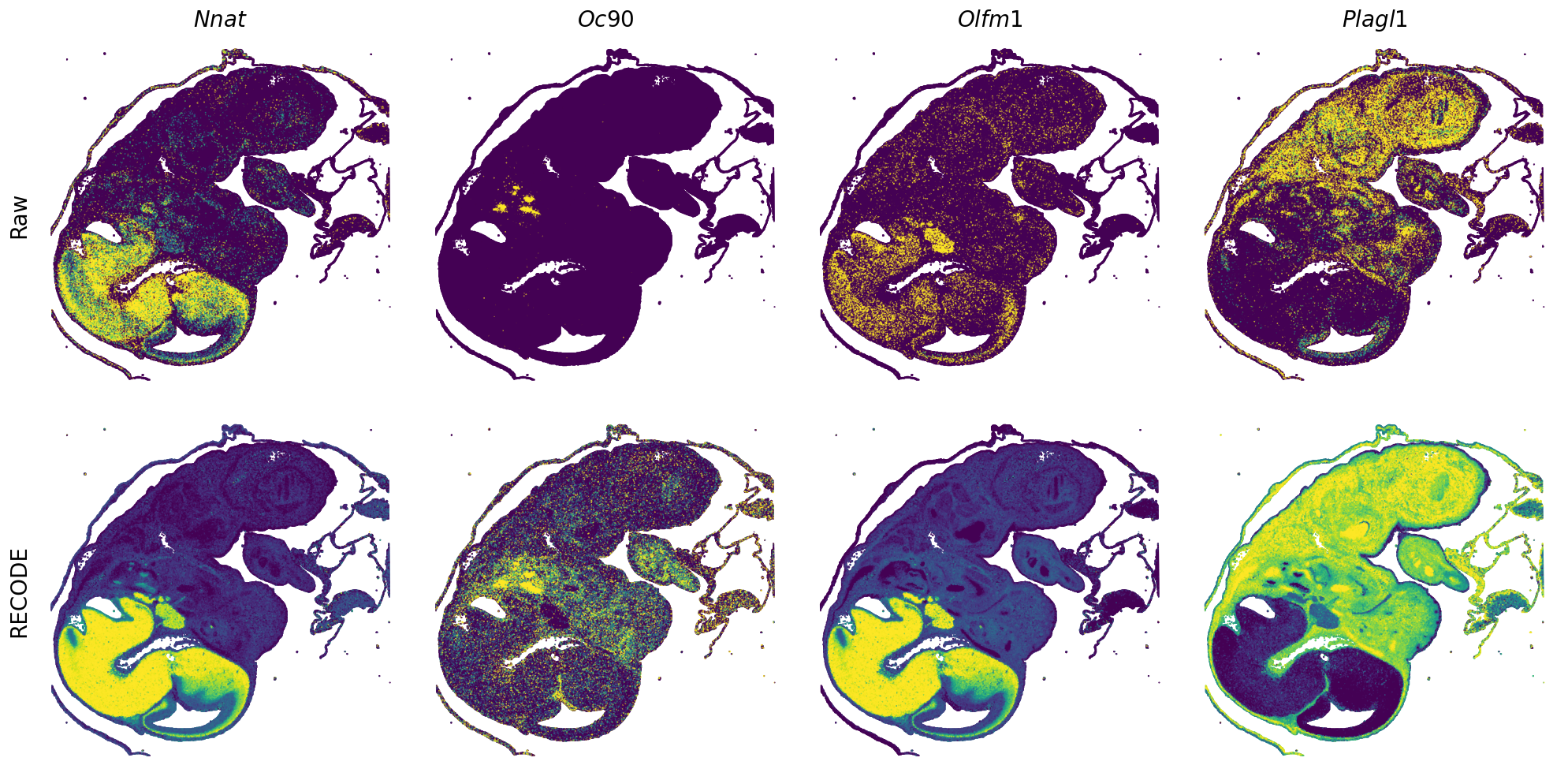
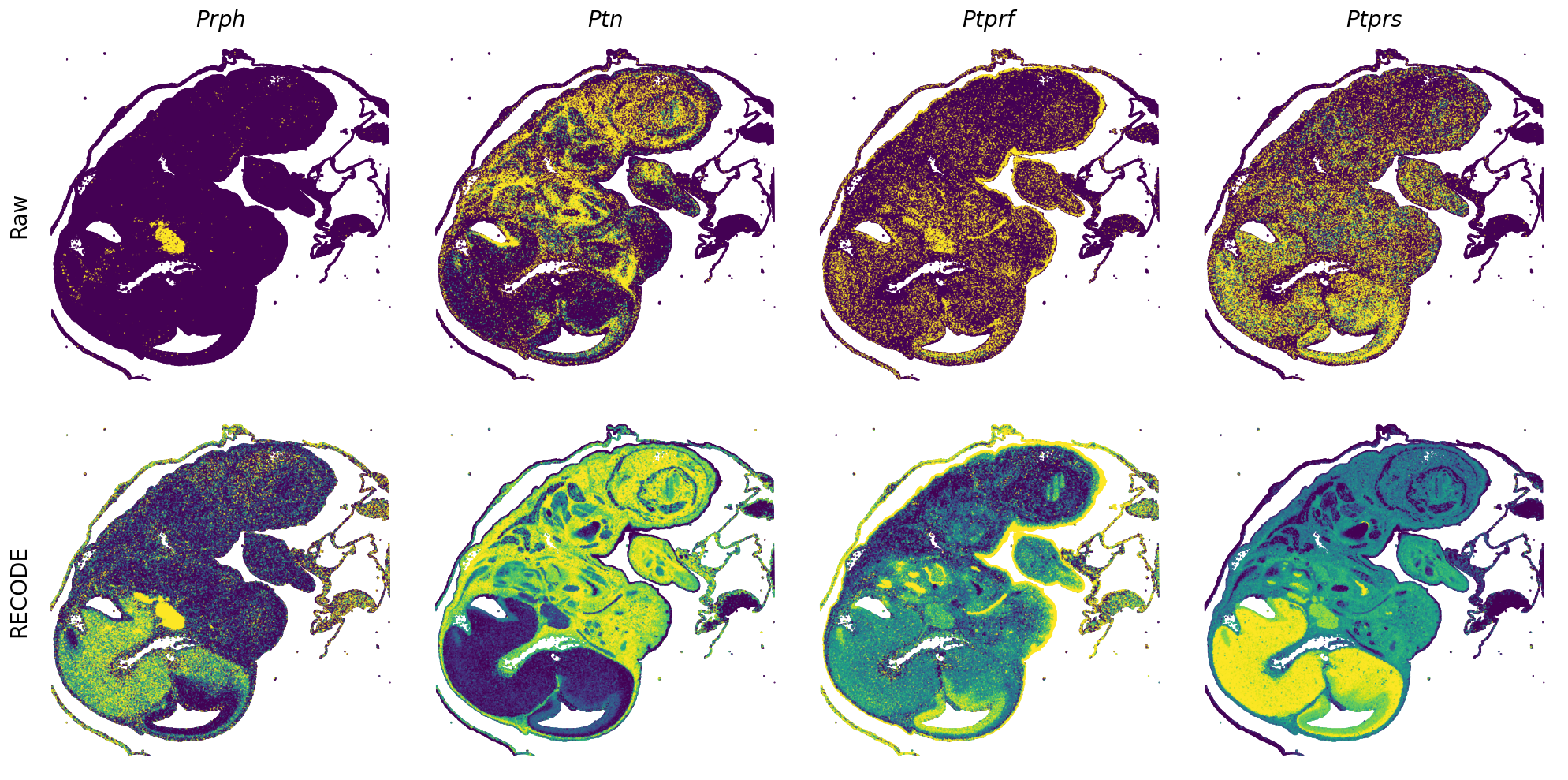
GENES = ["Acan", "Acta1", "Actc1", "Actn2", "Afp", "Agrn", "Ahnak", "Akap12", "Alb", "Aldoc", "Apoa1", "Apoa2", "Apoa4", "Apob", "Apoc2", "Apoe",
"App", "Cacng4", "Calm3", "Cd24a", "Cdkn1c", "Cenpb", "Clic6", "Cnmd", "Col11a2", "Col1a1", "Col1a2", "Col2a1", "Col3a1", "Col4a1", "Col9a1",
"Col9a2", "Col9a3", "Crabp1", "Crmp1", "Cubn", "Cxcl14", "Dcn", "Dcx", "Ddx17", "Dlk1", "Dsp", "Dynll2", "Fabp5", "Fabp7", "Fga", "Fgb",
"Fn1", "Gpx3", "Grb10", "H1f0", "H2afv", "Hapln1", "Hba-a2", "Hbb-bs", "Hbb-bt", "Hbb-y", "Hist1h1e", "Hist1h2ap", "Ibsp", "Igfbp2", "Igfbp4",
"Igfbp5", "Ina", "Itm2a", "Kctd12", "Kif1a", "Krt14", "Krt19", "Krt5", "Krt8", "Lrp2", "Lum", "Map1b", "Marcks", "Marcksl1", "Matn1", "Mest",
"mt-Co1", "mt-Nd1", "Myh8", "Mylpf", "Nefl", "Nefm", "Nnat", "Oc90", "Olfm1", "Plagl1", "Prph", "Ptn", "Ptprf", "Ptprs", "Pvalb", "Qk", "Rtn1",
"S100a11", "S100g", "Sbk1", "Serpinh1", "Slc6a15", "Sncg", "Sox11", "Sparc", "Spink1", "Spp2", "Stmn2", "Tac2", "Tceal7", "Thbs1", "Tnc", "Tnnc1",
"Tnnc2", "Tnni2", "Tnnt1", "Tnnt2", "Tnnt3", "Trp53inp2", "Ttn", "Ttr", "Tubb3", "Uchl1", "Vat1l", "Vim", "Wwp2"]
n_plots = 4
for i in range(int(len(GENES)/n_plots+0.9)):
if (i+1)*n_plots < len(GENES):
genes = GENES[i*n_plots:(i+1)*n_plots]
else:
genes = GENES[i*n_plots:len(GENES)]
spatial_gex(genes = genes)
[ ]: